eISSN: 2378-315X


Influenza A is the most important infectious disease confronting the world caused by Influenza A virus. It generally causes seasonal epidemic and pandemic flu with the high morbidity and mortality rate worldwide, mainly because the reservoir is difficult to eradicate as well as the virus, like other RNA viruses, is capable to undergo a continuous antigenic variation either via a point mutation in the surface antigens (called Antigenic drift) leading to seasonal epidemics or through a genetic reassortment (called Antigenic shift) which is a major change in the surface antigens leading to generation of entirely novel strains and sporadic pandemics among the population with no pre-existing immunity. Several targets such as (HA, NA, M2) are studied to inhibit binding and entry of virus into the target cells and prevent infection but due to high antigenic variations at these targets and the presence of drug resistance, it is found to be difficult to control the infection, therefore; alternative targets are thought to be crucial for treatment and management of potential future pandemics. Recently Nuclear export of NP are described as a common target of viral lifecycle for antiviral compounds. Some molecules such as RK424 and DP2392-E10 are found to inhibit the transportation of viral NP into- and from cytoplasm of infected cells and prevent the generation of viral progenies.
The purpose of this review is to describe the mechanism of seasonal epidemic and pandemic flu among the human population as well as to describe the recent targets of viral lifecycle for antiviral compounds to control and treat the unpredictable future pandemic infections.
Keywords: epidemic and pandemic flu, antigenic shift and drift, novel targets for anti-influenza compounds, NP antiviral compounds
HA, hemagglutinin; NA, neuraminidase; M2, membrane protein (Ione Channel); NP, nucleoprotein; vRNPs, viral ribonucleoproteins; CRM1, chromosome region membrane 1; NES3, nuclear export signal 3
Influenza or flu is a contagious respiratory disease of birds, human& many other mammals and it is caused by influenza viruses. The flu viruses are of three types A, B and C but only type A is mainly found to cause severe epidemics and pandemics among human population. This is because the influenza A virus is serologically having several different subtypes based on combination between their surface antigens known as Hemagglutinin (HA) and Neuraminidase (NA), which are 18 and 11 in number respectively.1,2 This serological variation is thought to enable the virus to undergo a constant antigenic variation to evade host immune system by using different mechanisms and as a result new strains with partially or completely changed surface antigens will develop which cannot be recognized by the pre-existing immunity against the older strains, either leading to a seasonal epidemic or spread more extensively causing a rare pandemic.1 Different kind of drugs and compounds with antiviral activities have been described at different stages and targets of viral life cycle, especially at the attachment and entry targets (e.g. HA, NA, and M2 inhibitors). This is; however, the emergence of drug resistance has been reported due to continuous antigenic variations at those targets. Therefore, other alternative targets of viral cycle are thought to be essential to control the viral replication and pandemic infections, and more recently nuclear export of NP is found to be an effective and alternative target for development of anti-influenza a compounds. Several compounds are described to be effective against this target and most common examples are leptomycin B, Verdinixor, RK424, DP2392-E10…etc.
Influenza a virus
Influenza A virus is an enveloped RNA virus that has two major surface proteins known as Hemagglutinin (HA) and neuraminidase (NA)3 The HA is a molecule that responsible for binding to specific receptors on the host cell surface where by which the virus picks up and produces infection whilst neuraminidase molecule is an enzyme that releases new virus particles produced in the infected cell through cleavage of sialic acid receptors from the cell surface and virion envelope to prevent aggregation of emerging viral particles.4,5 Influenza A viruses are reported to have 18 different HA molecules (H1-18) and 11 different NA molecules (NA1-11) on their cell surfaces till now (Table 1).2 Based on the combination of these two proteins, many different subtypes can be produced and each with a specific immunity.1 All influenza A strains are basically found in birds (with exception of H17N10 and H18N11which have only detected in bats)2,6–7 and only H1N1, H1N2 and H3N2 has described to infect human and pass person to person (Table 1). In addition, there are some other strains of the avian influenza A viruses can human infection such as H5N1, H7N2, H7N3, H7N7, H9, H7N9, H17N10 H18N112,8,9 but they are still not found to transmit among human populations and to cause pandemics.8
H Subtypes |
Human |
Bird |
Swine |
Bat |
N Subtypes |
Human |
Bird |
Swine |
Bat |
H 1 |
1934 |
1976 |
1930 |
N 1 |
1934 |
1959 |
1930 |
||
H 2 |
1957 |
1973 |
N 2 |
1957 |
1965 |
1970 |
|||
H 3 |
1968 |
1963 |
1970 |
N 3 |
1963 |
||||
H 4 |
1956 |
N 4 |
1968 |
||||||
H 5 |
1997 |
1961 |
N 5 |
1972 |
|||||
H 6 |
1965 |
N 6 |
1956 |
||||||
H 7 |
1996 |
1927 |
N 7 |
2003 |
1927 |
||||
H 8 |
1968 |
N 8 |
1963 |
||||||
H 9 |
1999 |
1966 |
N 9 |
2013 |
1974 |
||||
H 10 |
1949 |
N10 |
2009 |
|
2009 | ||||
H 11 |
1956 |
N11 |
2010 |
2010 |
|||||
H 12 |
1976 |
||||||||
H 13 |
1977 |
||||||||
H 14 |
1982 |
||||||||
H 15 |
1983 |
||||||||
H 16 |
2005 |
||||||||
H17 |
2009 |
2009 |
|||||||
H18 |
2010 |
|
2010 |
Epidemic and pandemic flu
Epidemic flu is a seasonal outbreak that attacks many people at about the same time and may spread through one or several countries and it occurs due to re-emergence of the previous year’s virus strain with a slight change in its surface binding proteins that may not be recognizable by the body's immune system. Whereas, the pandemic is found to be a matter of great concern and can cause a severe, easily transmissible disease among the community. It is a worldwide outbreak that caused by the emergence of new influenza A virus strain that can pass easily from person to person with no pre-existing immunity and everyone at risk of infection.10 By far four major influenza pandemics occur globally in the 20th and the 21st centuries which are Spanish flu in the 1918/1919, 'Asian flu' in the 1957/1958, 'Hong Kong flu' in the 1968/1969 and 'Pandemic (H1N1)' in (2009-2010) and each caused by different strains of influenza A. The deadliest pandemic was the Spanish flu outbreak, which had caused the highest morbidity and mortality rate among the population, probably more than 20 million people had died globally.5,11 This is increasing the risk that other pandemics may occur in the future either though the reintroduction of older avian strains (especially H3, H5 and H7 that are now circulating among the population) or by the generation of another new human strains.12 Therefore, the well understanding of influenza A strains and the process of them antigenic variation is essential and would be useful in dealing with any other possible epidemics and pandemics in the future.
Mechanism of influenza a virus antigenic variation
The mechanisms of surface antigenic variation by the influenza a viruses are commonly known as “antigenic drift” and “antigenic shift”. Antigenic drift is typically defined as a small change in the HA antigens caused by point mutation and immune selection pressure that directly leads to change of the HA epitopes (Figure 1). As it is well known, epitopes are binding sites of antigen that can be recognized by the components of host immune system, especially neutralizing antibodies. Therefore, any changes happen to these epitopes may not be recognition by the pre-existing antibody i.e. the new variant will replace the older ones and it may no longer or less effectively be recognized by the host immune system. As a result, it rapidly spread through the same population that had developed immunity to the previous strain leading to an epidemic arise. The epidemic outbreak occurs every few years and vaccines are updated every year based on strains projected to be circulating to minimize the infection. However, some people may still normally produce immunity against newly produced viruses because they still have some similarity to the older strains.13–15 While antigenic shift is generally described as a process of a sudden, major change of influenza A surface proteins (mainly HA) resulting from the recombination of the viral genomes of two different strains which may lead to introduction of entirely new strain into the human population with little or no immunity to them (Figure 1). So that everyone is at risk of acquiring infection and extensive epidemics or pandemics will result. The source of these new strains is primarily avian influenza viruses that found naturally in migratory wild birds (the natural reservoir).1,13–15
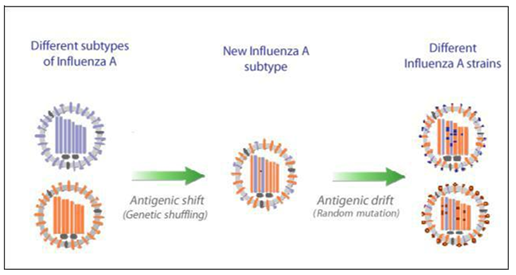
Figure 1This is a conceptual illustration of antigenic drift and antigenic shift. Antigenic drift produces viruses with a slightly-modified antigen, which is shown here as a change in a HA and NA molecules whilst antigenic shift generates viruses with entirely novel antigens, as shown here by recombination of viral segments of different subtypes. The figure is modified from.23
Generally, antigenic shift is described to occur through several ways: genetic reassortment; direct transmission and adaptation; re-introduction of old strains (Figure 2).
Genetic reassortment occurs through cross linking between two different strains of influenza A viruses in a single host cell (Figure 1), where the RNA segments from the two strains are mixed together during the new virus particles reassembly. As a result, a third viral strain will arise which contains the combined genes of both strains.13 Pigs and humans possess both receptors of human and avian influenza virus strains so that they can be infected by both strains at the same time leading to the mixed gene strains to emerge. 'Asian flu' in the 1957/1958, 'Hong Kong flu' in the 1968/1969 and Pandemic (H1N1) in (2009-2010) are good examples that are thought to be caused by genetic reassortment.
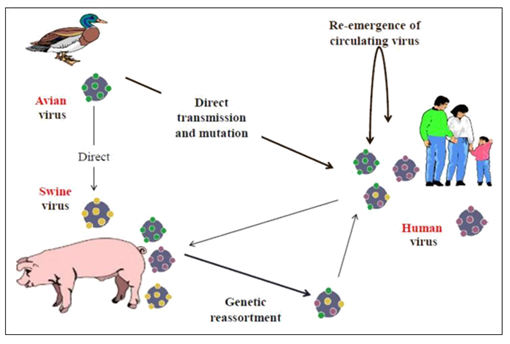
Figure 2 Demonstrates the concept of antigenic shift through three mechanisms: Genetic reassortment, direct transmission and Re-emergence.
Direct transmission and adaptation: The direct transmission of avian influenza A, especially with a highly pathogenic strain, has found to occur from birds to humans and then by adaptation of the virus in humans. This adaptation has occurred through changing the receptor binding site of HA from α 2, 3 linked sialic acid receptors in avian cells into α 2, 6 linked sialic acid receptors in human cells by mutation. As a result, a novel human strain might generate and be capable to transmit among the population and causing a pandemic. The ''Spanish flu'' in 1918/1919 is an example of pandemics that was thought to be transmitted by direct contact from avian to human beings based on the characteristic features of genetic material isolated from victims.
Re-emergence of old strains: Once the population’s immunity becomes weakened to a specific stain, then the virus can re-emerge and causing a pandemic outbreak among that population. For example, the Russian flu in 1950 and 1977 is found that it was probably caused by the re-emergence of the same H1N1 virus subtype.11–13
Antiviral drugs and targets
In recent decades, the development of antiviral medications is thought to be a crucial strategy in the management of seasonal epidemic and pandemic flu infections rather than the vaccination process because the development of vaccination against influenza A subtypes is thought to be insufficient and cost effective due to the presence of unexpected antigenic variation in influenza virus strains, the vaccine might require to be updated annually containing the new strains. Furthermore, it is also found that sometimes might produce ineffective immunity in immunocompromised patients.
Different kind of drugs and compounds are introduced and applied successfully. All are generally designed to interfere the viral replication cycle and inhibit cell infection through the targets in the viral life-cycle (Figure 3). At the beginning, antiviral medications are particularly focused on the impairment of the entry and attachment process of the virus to the host cell, such as HA, NA and M2 inhibitors, and then later other targets, such as RNA polymerase and endonuclease, are also involved in antiviral drug designing.16,17 Nafamostat, aprotinin, gabexate are examples of HA surface antigen inhibitors; Stachyflin, benzoquinones, BMY-27709 are also inhibitors of fusion and entry; zanamivir and oseltamivir are common examples of NA inhibitory drugs; amantadine and rimantadine are M2 ion channel blockers, although they are generally not recommended for treating influenza virus infections due to drug resistance concern; T-705, L-735, flutimide and their analogues are inhibitors of viral RNA polymerase and endonuclease.18,19

Figure 3 Illustrates the influenza A virus life cycle with recent and classical antiviral targets. The figure is modified from.24
Recent antiviral targets
After viral transcription and replication of the influenza RNA genome in the nucleus, the viral ribonucleoproteins (vRNPs) are exported into cytoplasm for the process of viral assembly and releasing. This export is reported that to be mediated by the cellular chromosome region maintenance 1 (CRM1) pathway, by the assistance of viral matrix protein 1(M1) and nuclear export proteins-NEP (NS2). NP directly binds to CRM1 and the interaction between NP/CRM1 and a recent identified host factor, nuclear export protein 1 (NXT1), promotes nuclear export of NP and vRNA.17,20
This viral nucleoprotein- mediated CRM1 are more recently become the main target of a variety of novel chemical compounds with anti-influenza activity; this is because NP plays an important role in the viral life cycle including vRNP nuclear transport (import and export) as well as transcription with replication of the viral genome. Research studies found that the inhibition of vRNP nuclear export might be a novel target for the development of anti-influenza compounds and be an effective strategy for impair influenza replication.17,21 Some anti-influenza molecules with NP-CRM1 inhibitory activity are described to potentially reduce replication of several influenza virus subtypes, such as RK424 and DP2392-E10, that they might interfere the nuclear export functions of NP and NEP via direct inhibition of the CRM1-NP and CRM1-NEP interactions, which are important for the process of production of new viral progenies.17,22–24
It is clear that influenza A virus is a real threat to human health, mostly due to the presence of non-eradicable reservoirs among migratory birds and the continuous capability of influenza A virus to undergo antigenic variation to evade the host immune system either through a slight gradual change in the surface glycoprotein's of the previous subtype (antigenic drift) causing seasonal epidemics or by a sudden big change in surface antigens (antigenic shift) with the emergence of entirely new strain that can cause serious illness and spread quickly from human-to-human resulting a rare pandemics. This is in case, the development of antiviral drugs is progressed but due to drug resistance and high antigenic variation at entry and attachment stages of viral replication cycle, the development of novel antiviral compounds and alternative targets is found to be more essential to control and manage the predictable future pandemic infections. Nuclear export of NP is recently become the common target for antiviral molecules and several compounds are found to target this part of viral replication lifecycle, especially DP2392-E10 compound which found can diminish viral replication of a broad range of influenza A subtypes by inhibiting nuclear export of viral NP and NEP as well as prevents the NP/CRM1 interaction by directly targeting the CRM1 protein.
Special thanks to the CAS-TWAS fellowships.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
2 7