eISSN: 2378-315X


Introduction: The single–nucleotide polymorphism (SNP) rs738409 C>G is located in the coding region of the patatin–like phospholipase domain–containing protein 3 (PNPLA3). It has been shown to be strongly associated with increased serum activities of transaminase enzymes, with risk of progression to cirrhosis and with fat accumulation in the liver. The study tests whether PNPLA3 rs738409 modifies the associations of body mass index (BMI) and alcohol consumption with serum transaminase levels (i.e., alanine transaminase (ALT) and aspartate transaminase (AST)).
Methods: Population–based data were drawn from two independent population–based cohorts within the Study of Health of Pomerania project (N= 8,056), conducted in Northeast Germany.
Results: We found several gene–environment interactions: The positive association between ALT and BMI was steeper for minor homozygotes (GG) and heterozygotes (CG) than for major homozygotes (CC) (p< 0.001). Similarly the relationship between AST and BMI in women was stronger with each G allele (p= 0.006 for CG, p= 0.001 for GG); higher levels of alcohol consumption were associated with steeper increase of AST activities for heterozygous men (p= 0.048).
Discussion: Results show that the SNP rs738409 modifies the associations of BMI and alcohol consumption with liver enzymes. This would have implications for guidance of patients if their genetic predisposition would be known. Heterozygous and especially minor homozygous subjects are inherently at particular high risk for liver–related problems. In addition, their risk rises even stronger in the presence of modifiable risk factors such as BMI or alcohol consumption.
Keywords: Gene–environment interactions; Study of Health in Pomerania
SNP, single–nucleotide polymorphism; ALT, alanine transaminase; AST, aspartate transaminase; GG, minor homozygotes; CG, heterozygotes; CC, major homozygotes; BMI, body mass index; NAFLD, non–alcoholic fatty liver disease; SHIP, study of health in pomerania
For the common rs738409 C>G SNP, which is located in the patatin–like phospholipase domain–containing protein 3 (PNPLA3), a strong association between non–alcoholic fatty liver disease (NAFLD) has been detected,1 approved2–8 and expanded to an association with liver–fat content in general8–13 and serum transaminase levels.14 Also a association to a higher risk of progression to cirrhosis in NAFLD patients has been shown.15 Transaminase levels, particularly of alanine transaminase (ALT) and aspartate transaminase (AST), are strong indicators of hepatic steatosis16 as well as of cardio–metabolic risks17 and are associated with mortality even within their reference ranges.18 Yet, a small number of studies reported gene–environment interactions for rs738409. Recently it has been shown that the G allele of rs738409 is associated with lower serum triglyceride and cholesterol levels and higher ALT levels in overweight individuals only.19 Children with the minor G allele had more liver fat per unit omega–fat intake than major homozygous children.20 In another study, sugar and carbohydrate intake in Hispanic children was positively related to hepatic fat fraction in GG carriers but not in CG and CC carriers.21
Because both the genetic variant and environmental conditions such as physical activity or obesity influence serum–transaminase levels,22 we consider them as a valuable model for the examination of gene–environment interactions. We chose body mass index (BMI) and alcohol consumption as common environmental factors because they are tightly associated with liver–enzyme activity,23 liver–fat content, hepatic steatosis and increased risk of liver fibrosis and cirrhosis.24 Further, they may accelerate insulin resistance and cardiovascular diseases25 and are therefore highly relevant for health care. In addition, BMI and alcohol consumption can be in general modified by changing the individual’s life style. Regarding the consequences of different genetic makeups, physicians and patients would like to know: Is the genetic risk associated with patient related factors that can be influenced by medical treatment or behavioural changes? The study tests whether PNPLA3 rs738409 modifies the associations of body mass index (BMI) and alcohol consumption with serum transaminase levels (i.e., alanine transaminase (ALT), aspartate transaminase (AST)).
Population and study design
To investigate the relationship between rs738409, transaminase levels, BMI and alcohol consumption, we used data from the baseline Study of Health in Pomerania (SHIP).26,27 The study population consists of adult Caucasian residents of Western Pomerania in Northeast Germany. From the total population of West Pomerania comprising 213,057 inhabitants in 1996, a two–stage stratified random cluster sample of adults aged 20–79 years was drawn (for further details see).26 The net sample comprised 6265 eligible subjects. Of these, 4308 subjects (n= 2192 women; response of 68.8%) participated in the SHIP–0 between 1997 and 2001. A separate stratified random sample of 8016 adults aged 20–79 years was drawn for the SHIP–TREND cohort between 2008 and 2011 of which 4420 participated (N=2274 women, response of 55.1%). We pooled both cohorts for this analysis.
Out of the 8728 SHIP subjects, genotypes were missing in 375 (4.3%) of the individuals due to insufficient imputation quality (call rate<86 %). Missing genotype information was not significantly associated with and ALT and AST activities (logistic regression, adjusted for age and sex, with missed outcome, p=0.096, N=8689 and p=0.610, N=8694, respectively). For another 297 individuals (3.4%) covariates were missing, in 255 datasets concerning alcohol consumption. Thus, the analytic sample comprised 8056 participants aged 20–83 years (N=4134 women). Approval was obtained by the local ethic committee, and informed consent was given by all participants.
Genotyping and imputation
DNA samples of were genotyped using the Affymetrix Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA), genotypes were determined using Birdseed2 clustering algorithm.27 Imputation of the SNP rs738409 was performed with the software IMPUTE against HapMap II CEU. Details of the genotyping and imputation are reported elsewhere.28 Instead of the raw imputed values, we used rounded values from the imputed SNP (0,1,2). Thus we used genotype as a categorical variable resulting in a codominant genetic model that is most powerful to detect associations when the inheritance model is not known.29 Arrays with a QC–call rate < 86% were excluded. The final call rate for genotyping was>0.99 for the SHIP–0 and>0.92 for the SHIP–TREND. Genetic data were stored using the database Cache (Inter Systems, Cambridge, MA, USA).
Data collection
Data were gathered by trained and certified medical staff. Self–reported alcohol consumption was assessed using a beverage–specific quantity–frequency measure:30 number of days with alcohol consumption (beer, wine, spirits) and average daily alcohol consumption for such a day over the past month. Average alcohol consumption (in grams per day) was calculated by multiplying frequency and amount of alcohol from beer, wine and spirits, respectively, using a standard ethanol content of 4.8 percent (by volume) in beer, 11 percent in wine, and 33 percent in spirits to conversion.31 BMI was calculated as kg/m2. Non–fasting blood samples were drawn from the cubital vein in the supine position. Serum ALT and AST were measured photometrically at 37 °C (Hitachi 717 and 704, Roche, Mannheim, Germany). Further details on the study protocol can be found in.28
Statistical analyses
We applied ordinary least–square linear regression; since the relationships between liver enzymes and covariates were expected to be non–linear, we transformed the independent variables if appropriate to get a linear relationship.32 We modelled gene–environment interactions with the STATA program ‘mfpigen’ with an alpha level for interactions of 0.05.33,34 The polynomial to be fit was restricted to two degrees to avoid overfitting. Since variances were heteroscedastic, we applied robust standard errors. Models were adjusted for age and sex, an approach suitable in population–based studies.35 Statistical analyses were performed using STATA 13.0 (Stata Corp., College Station, TX, USA). For the display of the interaction effects we used predictive margins with age fixed at 50 years and 50% females for the unisex model.
In higher ranges of alcohol consumption data were very sparse. We therefore restricted the analyses to the range with sufficient data and eliminated for these analyses datasets with more than 60 g alcohol per day in women (N= 6) and more than 130 g in men (N= 4), additionally with more than 13 g in minor homozygous women (N= 15) and with more than 60 g in minor homozygous men (N= 7).
For models examining gene–environment interactions also interactions with all covariates should be included to regard effect modifications of possible confounders.36 For example, if we model an interaction effect between alcohol consumption and a genetic group, adjusting for age and sex, a model should include also the interactions genetic group × age and genetic group × sex. However, since we considered a confounding effect of age and sex as unlikely, we conducted the main analyses without these interaction terms. As a sensitivity analysis we included all interactions between covariates and SNP in the models which did not considerably change the results.
In our study population of 8056 individuals, 60.4% were major homozygous (CC), 34.7% heterozygous (CG) and 4.9% minor homozygous (GG; Table 1). Age and sex were not significantly distributed among genetic groups with a slightly higher proportion of women in the minor homozygous group (55.0% compared to overall 51.3%). Carriers of the G allele of PNPLA3 rs73809 did not significantly differ regarding BMI (p = 0.887) and alcohol consumption (p= 0.298) but had increased liver enzymes compared to CC subjects (p < 0.001, Table 1).
Characteristic |
Variant of Rs738409 |
p Value |
||
|---|---|---|---|---|
CC |
CG |
GG |
||
rs738409 (%) |
4865 (60.4%) |
2798 (34.7%) |
393 (4.9%) |
|
Age (years)2 adj for sex |
50.9 (15.8) |
50.4 (16.0) |
51.3 (15.5) |
0.328 |
Female sex (%)1 adj for age |
2457 (50.5%) |
1461 (52.2%) |
216 (55.0%) |
0.123 |
Body Mass Index (kg/m2)* |
28.3 (28.13, 28.47) |
28.24 (28.03, 28.46) |
28.25 (27.76, 28.74) |
0.887 |
Alcohol Consumption (g/d)* |
11.8 (11.2, 12.3) |
11.9 (11.2, 12.5) |
10.7 (9.3, 12.1) |
0.298 |
Alanine Transaminase (µmol ∙ sl−1)* |
0.491 (0.480, 0.501) |
0.532 (0.517, 0.547) |
0.607 (0.567, 0.646) |
< 0.001 |
Aspartate Transaminase (µmol ∙ sl−1)* |
0.349 (0.343, 0.356) |
0.372 (0.362, 0.383) |
0.419 (0.390, 0.447) |
< 0.001 |
Table 1 Characteristics of the study population
Age is given as marginal mean adjusted for sex with 50% women (standard deviation); Rs738409 is given as numbers (%); Sex is given as marginal mean adjusted for age; Body mass index, Liver enzymes and alcohol consumption are given as marignal means fixed at an age of 50 years and 50% women (95% confidence interval); p values refer to a combined test over all three genotypes.
Relationship of BMI and alcohol consumption with transaminase levels were modified by PNPLA3 rs73809. The association between of BMI and ALT got stronger with each additional G allele (p < 0.001, Table 2, Figure 1 A). For all other relationships, a significant interaction with sex was detected (at least one of CG or GG with a p value < 0.05 for the three–fold interaction SNP × sex × alcohol, not shown). Consequently, further analyses were conducted separately for men and women. There was no significant interaction between the SNP and alcohol consumption with ALT used as outcome. (Table 2)
Coefficient |
p |
95% Confidence |
Interval |
|
Men and women (N = 8056, R2 = 0.167) |
||||
rs738409 |
||||
CC |
Reference |
|||
GC |
0.0471 |
< 0.001 |
0.0318 |
0.0625 |
GG |
0.1323 |
< 0.001 |
0.0927 |
0.1718 |
BMI (kg/m2; Polynomial: BMI-1) |
-1.075 |
< 0.001 |
-1.195 |
-0.9551 |
Age (Years; Polynomial: age3) |
-0.0004 |
< 0.001 |
-0.0004 |
-0.0003 |
Sex (Female) |
-0.1776 |
< 0.001 |
-0.1906 |
-0.1647 |
rs738409 x BMI |
||||
CC |
Reference |
|||
GC |
-0.3663 |
< 0.001 |
-0.5661 |
-0.1665 |
GG |
-1.1166 |
< 0.001 |
-1.6412 |
-0.5921 |
Men only (N = 3914, R2 = 0.070) |
||||
rs738409 |
||||
CC |
Reference |
|||
GC |
0.0613 |
< 0.001 |
0.0359 |
0.0867 |
GG |
0.1702 |
< 0.001 |
0.1035 |
0.2369 |
Alcohol Consumption (g/day) |
0.0019 |
< 0.001 |
0.0011 |
0.0027 |
Age (years) |
-0.0005 |
< 0.001 |
-0.0006 |
-0.0004 |
rs738409 x alcohol |
||||
CC |
Reference |
|||
GC |
0.001 |
0.196 |
-0.0005 |
0.0025 |
GG |
0.0031 |
0.193 |
-0.0016 |
0.0077 |
Women only (N= 4119, R2 = 0.030) |
||||
rs738409 |
||||
CC |
Reference |
|||
GC |
0.0145 |
0.053 |
-0.0016 |
0.0291 |
GG |
0.0693 |
< 0.001 |
0.0304 |
0.1083 |
Alcohol Consumption (g/day) |
0.0008 |
0.46 |
-0.0013 |
0.003 |
Age (years; polynomial: age-0.5) |
-0.4097 |
< 0.001 |
-0.485 |
-0.3344 |
rs738409 x alcohol |
||||
CC |
Reference |
|||
GC |
-0.0013 |
0.357 |
-0.004 |
0.0014 |
GG |
-0.0099 |
0.184 |
-0.0245 |
0.0047 |
Table 2 Results from regression with ALT as outcome and interaction between body mass index/alcohol consumption and rs738409
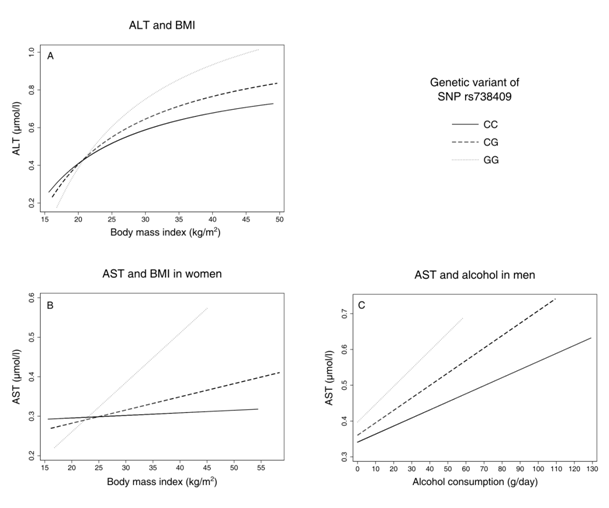
Figure 1 Predictive margins between liver-enzyme activities and environmental factors body mass index (BMI) and alcohol consumption by genetic group, adjusted for age (all models) and sex (first model, panel A); ALT, Alanine Transaminase; AST, Aspartate Transaminase.
Regarding AST and BMI, there was no significant interaction in men (p= 0.737 for CG, p= 0.197 for GG) but in women (p= 0.006 for CG, p= 0.001 for GG, Table 3, Figure 1B). In men, higher levels of alcohol consumption were associated with slightly higher AST values for each G allele, being significant for GC (p= 0.048 for CG, p= 0.386 for GG, Table 4, Figure 1). In women, there was no significant interaction between the genetic variant and alcohol consumption (p= 0.338 for CG, p= 0.121 for GG, Table 4).
Coefficient |
p |
95% Confidence |
Interval |
|
|---|---|---|---|---|
Men only (N = 3914, R2 = 0.050) |
||||
rs738409 |
||||
CC |
Reference |
|||
GC |
0.0375 |
< 0.001 |
0.0196 |
0.0554 |
GG |
0.0968 |
0.001 |
0.0401 |
0.1534 |
Alcohol consumption |
0.0023 |
< 0.001 |
0.0017 |
0.0028 |
Age (years) |
-0.0002 |
0.289 |
-0.0007 |
0.0002 |
rs738409 x alcohol |
||||
CC |
Reference |
|||
GC |
0.0012 |
0.048 |
0.00001 |
0.0024 |
GG |
0.0027 |
0.386 |
-0.0034 |
0.0089 |
Women only (N = 4107, R2 = 0.043) |
||||
rs738409 |
||||
CC |
Reference |
|||
GC |
0.0081 |
0.08 |
-0.001 |
0.0171 |
GG |
0.0586 |
0.002 |
0.0207 |
0.0966 |
Alcohol Consumption (g/day, Polynomial: Alcohol3) |
0.0017 |
0.086 |
-0.0002 |
0.0036 |
Age (Years) |
0.0016 |
0 |
0.0014 |
0.0019 |
rs738409 x alcohol |
||||
CC |
Reference |
|||
GC |
-0.001 |
0.338 |
-0.0029 |
0.001 |
GG |
-0.013 |
0.121 |
-0.0293 |
0.0034 |
Table 3 Amplitude in mill volts of the Lead-1 of electrocardiography in sheep
*Significant (P≤0.05); NSNot significant (P>0.05)
Coefficient |
p |
95% Confidence |
Interval |
|
|---|---|---|---|---|
Men only (N = 3914, R2 = 0.050) |
||||
rs738409 |
||||
CC |
Reference |
|||
GC |
0.0375 |
< 0.001 |
0.0196 |
0.0554 |
GG |
0.0968 |
0.001 |
0.0401 |
0.1534 |
Alcohol consumption |
0.0023 |
< 0.001 |
0.0017 |
0.0028 |
Age (years) |
-0.0002 |
0.289 |
-0.0007 |
0.0002 |
rs738409 x alcohol |
||||
CC |
Reference |
|||
GC |
0.0012 |
0.048 |
0.00001 |
0.0024 |
GG |
0.0027 |
0.386 |
-0.0034 |
0.0089 |
Women only (N = 4107, R2 = 0.043) |
||||
rs738409 |
||||
CC |
Reference |
|||
GC |
0.0081 |
0.08 |
-0.001 |
0.0171 |
GG |
0.0586 |
0.002 |
0.0207 |
0.0966 |
Alcohol Consumption (g/day, Polynomial: Alcohol3) |
0.0017 |
0.086 |
-0.0002 |
0.0036 |
Age (Years) |
0.0016 |
0 |
0.0014 |
0.0019 |
rs738409 x alcohol |
||||
CC |
Reference |
|||
GC |
-0.001 |
0.338 |
-0.0029 |
0.001 |
GG |
-0.013 |
0.121 |
-0.0293 |
0.0034 |
Table 4 Results from regression with AST as outcome and interaction between alcohol consumption and rs738409
We found interactions between PNPLA3 rs73809 and BMI and alcohol consumption, respectively, that together influenced liver enzyme levels. The increase in ALT levels was steeper for each G allele in BMI (Figure 1A), similarly for AST in women (Figure 1B) and AST and alcohol in men. (Figure 1C) In all cases, effect sizes of GG on BMI and alcohol were more than twice as high as for CG (on average about fourfold), indicating a more than additive effect of the G alleles. Our results further show that there are sex–specific differences in SNP–related transaminase levels: The increase in transaminase levels per G allele is much steeper in men compared to women, about fivefold for CG and twofold for GG. This implies that, besides the effect of gene–environment interactions, women seem to be at lower risk for increased transaminase levels when carrying a G allele.
In line with our findings previous studies also found not significant differences in BMI between the genetic groups.37,38 Several studies report an association of the G allele of rs738409 with alcoholic liver disease and alcoholic cirrhosis.39–42 This finding is supported for men by our results; however, previous studies did not explicitly examine sex–specific differences for this association and examined mainly male alcoholics. Regarding gene–environment interactions, dietary issues matter as well in children20,21 as in adults for which it has been reported that with higher consumption of sucrose the increase in serum triglycerides was higher with each G allele.19 For ALT we confirmed previous findings19 that there are only differences in ALT levels between the genetic groups in overweight individuals. Our findings illustrate that gene–environment interactions may contribute to the heritability of liver–related problems. If these will be confirmed in further validation studies, this would suggest some implications for patients and clinicians. A physician who has genetic information on rs738409 available for a patient, he or she may warn patients with a G allele to be particularly cautious because their transaminase levels increase stronger with increasing BMI or alcohol consumption compared to CC individuals
The SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, 01ZZ0403 and 03IS2061A), the Ministry of Cultural Affairs as well as the Social Ministry of the Federal State of Mecklenburg–West Pomerania. We also thank Mr. Alexander Teumer for valuable contributions that improved the initial manuscript.
Author declares that their is no conflict.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
2 7