Advances in
eISSN: 2572-8490


Research Article Volume 1 Issue 1
1Biosystems and Biomaterials Division, National Institute of Standards & Technology, USA
2Department of Physics, University of Maryland, USA
Correspondence: Sarkar, National Institute of Standards and Technology, 100 Bureau Dr, Gaithersburg, MD20899, USA
Received: July 14, 2016 | Published: August 22, 2016
Citation: Sarkar S, Baker BA, Chen D, et al. Roles of nanofiber scaffold structure and chemistry in directing human bone marrow stromal cell response. Adv Tissue Eng Regen Med Open Access. 2016;1(1):6-18. DOI: 10.15406/atroa.2016.01.00003
Nanofiber technology has emerged as a promising tool to recapitulate native extracellular matrix structure; however the properties of nanofibers governing cell-material interactions are still largely undetermined. We have investigated the role of both nanofiber structure and chemistry in directing the response of human bone marrow stromal cells (hBMSCs). We developed a poly (ε-caprolactone) (PCL) nanofiber material system where scaffold structure is maintained while surface chemistry is modified via mild sodium hydroxide (NaOH) hydrolysis to introduce surface carboxyl groups. Surface carboxyl groups was verified via x-ray photoelectron spectroscopy (XPS), water contact angle (WCA), and toluidine blue-O biochemical assay and scaffold structure was verified with scanning electron microscopy (SEM). hBMSC response was investigated for proliferation, differentiation (alkaline phosphatase activity and mineralization) , cell morphology, and microarray gene expression profiles. We found that both nanofiber structure and chemistry play a role in hBMSC osteogenic differentiation where PCL nanofiber scaffolds were able to elicit an osteogenic phenotype, while chemically modified PCL nanofiber scaffolds did not. Scaffold structure was found to have a more dominant effect on cell morphology and extracellular matrix gene expression while nanofiber chemistry had a larger effect on genes related to metabolism and cytoskeletal rearrangement. The nanofiber scaffold material system developed here can serve as a platform to further investigate the role of micro environmental cues on osteogenic differentiation. The NaOH modified NF scaffolds are particularly useful as a negative control for osteogenic differentiation in NF scaffolds.
Keywords:nanofiber scaffold, stem cells, surface modification, bone tissue engineering, microarray gene expression, cell morphology
ECM, extracellular matrix; COOH, carboxyl functionality; NH2, amine functionality; OH, hydroxyl functionality; hBMSC, human bone marrow stromal cell; NF, pcl non-woven nanofibers; TCPS, tissue culture polystyrene; SEM, scanning electron microscopy; SC films, PCL spin-coated TCPS discs; NaOH, sodium hydroxide; DI, de ionized; MOD NF, nf scaffolds modified by mild hydrolysis; MOD SC, sc films modified by mild hydrolysis; TBO, toluidine blue-o; XPS, x-ray photoelectron spectroscopy; At %, atomic concentrations; CO2, carbon dioxide; EDTA, ethylene diamine tetra actate; OS, osteogenic supplement; PBS, phosphate buffered salt solution; DNA, deoxyribonucleic acid; ALP, alkaline phosphatase; AR, alizarin red; BSA, bovine serum albumin; NA, numerical aperture; RNA, ribonucleic acid; NIST, national institute of standards and technology; ERCC, external rna controls consortium; SAM, significance analysis of microarrays; ALL NF, NF and MOD NF; ALL SC, SC and MOD SC; ALL UN-MOD, NF and SC; GO, gene ontology; KEGG, kyoto encyclopedia of genes and genomes; DAVID, database for annotation visualization and integrated discovery; WCA, water contact angle; Cν, coefficient of variation; MC3T3-E1, osteoblast-like cells; ALPL gene for alkaline phosphatase; COL8A2, gene for Collagen Type VIII Alpha 2; FNDC1, gene for fibronectin type III domain containing 1, FBLN5, gene for fibulin 5; GPC4, gene for glypican 4; IGFBP1, gene for insulin like growth factor binding protein 1; IGF, insulin like growth factor; IL 8, gene for interleukin 8; KRT18, gene for keratin 18; RHOA, gene forras homolog gene family; TUBB6, gene for tubulin; TUBBP1, gene for similar to tubulin; coli, Escherichia coli; PFKFB4, gene for 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4; TPI1, gene for triosephosphate isomerase 1; ENO2, gene for enolase 2; PCK2, gene for phosphoenolpyruvate carboxykinase 2; PFKP, gene for phosphofructokinase
Nanofiber technology has emerged as a promising tool to recapitulate the native extracellular matrix (ECM) environment for tissue engineering and regenerative medicine strategies.1–3 A wide range of fibrous scaffolds have been fabricated from both natural and synthetic polymers with control over fiber diameter, alignment, chemical and biological functionalization, and scaffold properties.3–5 Nanofiber materials have been demonstrated to promote desirable cell-material interactions that affect properties such as cell attachment, morphology, differentiation, protein adsorption and ECM deposition.3
Structural cues in the nanofiber environment are often attributed to these effects and are largely dependent on the nanofiber mat properties such as fiber diameter, alignment and porosity.4,6–8 Biological functionalization and chemical modification have also been demonstrated have significant effects on NF scaffolds. For example, Chua et al.7 functionalized the surface of polyether sulfone nanofibers and investigated the effects of carboxyl (-COOH), amine (-NH2) and hydroxyl (-OH) functionalities on hematopoietic stem/progenitor cell culture expansion.7 They found that amine functionalized nanofibers supported the highest degree of cell adhesion and expansion however the mechanisms behind this enhanced functionality are largely undetermined.7 In order to better understand the mechanisms underlying cell-material interactions in the nanofiber environment, systematic studies with controlled material properties and culture conditions are required.
Several studies have demonstrated the strong potential for the use of nanofiber scaffolds in bone tissue engineering, where osteogenic differentiation is enhanced on nanofiber scaffolds.9–13 In a recent study conducted by Kumar et al.,14 the importance of the nanofiber scaffold structure in directing stem cell fate down an osteogenic linage was further supported. The study isolated the effect of scaffold structural cues in the direction of stem cell fate by systematically investigating scaffolds of similar chemical compositions with varying 3D architectures. Surprisingly, only nanofiber scaffolds were able to promote human bone marrow stromal cell (hBMSC) osteogenic differentiation in the absence of osteogenic differentiation supplements.14 Cells on these materials also had elongated branched morphologies and cytoskeletal gene expression profiles consistent with those cells cultured in the presence of osteogenic supplement, supporting possible cell morphology based mechanisms behind stem-cell nanofiber interactions.14 In the current study, we further investigate the role of cell morphology in the osteogenic response of stem cells to nanofiber scaffolds.
To investigate the contributions of both nanofiber structure and surface chemistry in directing stem cell response and osteogenic differentiation, we have developed a material system with nanofiber scaffolds of similar structure but significantly different chemistry. Methods are available to tune and modify the surface chemistry of nanofibers.8,15–17 However, surface modification of the nanofibers can be destructive to the nanofiber morphology. This is especially apparent in plasma oxidation of nanofiber scaffolds.6,18 To minimize physical variability in nanofiber mat structure while changing fiber chemistry, we have utilized the wet chemical surface modification technique of base hydrolysis.8,11,15,19–21 To minimally effect scaffold structure while significantly altering scaffold surface chemistry. hBMSC proliferation, osteogenic differentiation, morphology, and microarray gene expression were then investigated on this material system as a function of scaffold structure and chemistry to better understand the contributions of these properties on stem cell response to nanofiber materials.
Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
Electrospun nanofiber mat fabrication
Nanofibers were fabricated from poly (ε-caprolactone) (PCL, relative molecular mass 80,000 g/mL, Sigma) using a custom-built electrospinning apparatus.14 Electrospinning parameters such as solvent composition (3:1-9:1 volume ratio of chloroform: methanol), flow rate (0.5mL/h and 2mL/h), and PCL concentration (0.1g/mL and 0.15g/mL) were systematically varied (data not shown) to obtain fibers of approximately 600 nm diameter with low fiber diameter variability (Coefficient of variation (Cʋ) of 0.14). PCL solution was prepared in 5:1 volume ratio chloroform: methanol solution at a 0.15g/mL PCL concentration. The polymer solution was loaded into a 3mL syringe and dispensed with a syringe pump at 0.5mL/h through a 21 gauge 1" shaft, flat tip, dispensing needle. The positive lead from a power supply was affixed to the needle while the ground lead was fixed to the target (aluminum foil collection plate). The distance between the needle and target was approximately 21cm and the voltage applied was 13kV. Non-woven PCL nanofibers (NF) were collected onto approximately 0.95cm diameter hot punched tissue culture polystyrene (TCPS) discs for 6 h (3mL of solution dispensed). NFs collected on TCPS discs were removed from the foil target and assessed for morphology with scanning electron microscopy (SEM) or adhered to the bottom of 48-well plates using silicone grease (Dupont) for cell culture experiments.
2D spin coated PCL films
PCL coated surfaces were prepared by spin-coating PCL solution (0.7mL, 0.1g/mL PCL in glacial acetic acid) at 104.72 rad/s (1000 rpm) for 20s onto a TCPS dish (100 mm diameter). Films were air dried at room temperature overnight then heated above 60°C repeatedly to adhere to the PCL to the TCPS surface as well as achieve a characteristic and repeatable cobblestone pattern. PCL coated TCPS discs (SC films) 0.95 cm in diameter were then hot punched from the TCPS dish. SC films were prepared for SEM imaging and chemical characterization or adhered to the bottom of 48-well plates for cell culture. These SC films serve as a control 2-D analogues to the NF scaffolds.
Scaffold sterilization
NF scaffolds and SC films were secured in 48-well plates with silicone grease. Plates were then sterilized by ethylene oxide treatment (12 h cycle, 48 h degassing under house vacuum). All subsequent handling was carried out in sterile conditions.
Scaffold chemical modification
NF scaffolds and SC films were treated via mild hydrolysis in sodium hydroxide (NaOH) solution in order to expose carboxyl (COOH) and hydroxyl (OH) groups on the material surface and render scaffolds more hydrophilic (Figure 1).15,19 Scaffold hydrolysis was tuned to maximize surface COOH concentration while minimizing structural changes. Parameters including NaOH concentration (0.1mol/L to 5mol/L), incubation time (1h to 7h), and incubation temperature (25°C and 37°C) were systematically varied (Data not shown). In a final procedure, scaffolds were hydrolyzed in freshly prepared 1mol/L sodium hydroxide (NaOH) solution in sterile filtered de ionized (DI) water. Briefly, 500µL of 1 mol/L NaOH solution was pipetted onto the scaffolds (500µL of DI water was used for the un-modified case) and incubated at 37°C under constant agitation for 7h. Scaffolds were then rinsed 2x with sterile DI water followed by a 30 min rinse in DI water under constant agitation. Scaffolds were rinsed again 2x with sterile DI water, then set in a sterile cell culture hood overnight to dry. NF scaffolds and SC films modified by mild hydrolysis are denoted as MOD NF and MOD SC respectively. Modified scaffolds were tested immediately for surface chemical properties or immediately immersed in cell culture medium containing fetal bovine serum, in preparation for cell seeding.

Scanning electron microscopy (SEM)
NF and MOD NF scaffold and SC and MOD SC film morphology were investigated using SEM. Substrates were sputter coated with gold (90s, 15mA) then imaged on a Hitachi S-4700-II FE-SEM at 5kV (5000X and 50,000X magnification). Fiber diameter was determined from scanning electron micrographs from five fields of view using Image J software (National Institute of Health) to determine average fiber diameter and fiber diameter variability (>80 fibers investigated per group) before and after chemical modification.
Contact angle goniometry
Surface wettability was investigated via water contact angle goniometery. DI Water droplets (approximately 100µL) were dispensed onto the scaffold and film surfaces, imaged and modeled using sessile drop fitting to obtain contact angle measurement (averaged across 4 samples, 3 spots per sample).
Toluidine blue-o assay for carboxyl functional groups
Carboxyl concentration was were soaked in 1 ml of 0.5 mmol/L TBO (buffered to pH 10determined using the toluidine blue-O (TBO) reagent.22–26 Substrates at room temperature) for 5 h under constant agitation. Non-complexed dye was then removed by rinsing in an excess of 0.1 mmol/L NaOH. Complexed dye was then desorbed from the surface by incubating each substrate in 500µL of 500mL/L glacial acetic acid solution in DI for 10 min under vortexing. Desorbed dye solution was then plated into a 96-well plate (200µL per well) and the absorbance was measured at 633 nm wavelength. A calibration curve of TBO solutions of known TBO concentration was generated to determine the concentration of carboxyl groups on the scaffold or film surface.24,25 The surface density of carboxyl groups was calculated under the assumption that the TBO complexes to equivalent moles of carboxyl groups on the surface (a 1:1 carboxyl: TBO relationship).22,23
X-ray photoelectron spectroscopy (XPS)
XPS was used to conduct an elemental analysis of the NF, MOD NF, SC and MOD SC surfaces. Spectra were obtained on a Kratos AXIS Ultra DLD spectrometer with a monochromatic Al x-ray source (1486.7eV) operating at 140 W under approximately 1.0 x 10-9 Torr (1.33x10-11 Pa) vacuum (Supplemental Information M1). Atomic concentrations (At %) were calculated from survey spectra. Three spectra were acquired for each sample, and triplicate samples were investigated for each group.
Human bone marrow stromal cell (hBMSC) culture
Sterile TCPS discs, NF scaffolds, MOD NF scaffolds, SC films, and MOD SC films secured in 48-well plates were soaked in culture medium (α-minimum essential medium (In vitro gen) supplemented with 165mL/L fetal bovine serum (FBS, Atlanta Biologics), 4mmol/L ι-glutamine and 10mL/L Penicillin/Streptomycin (Cellgro)) for 48 h to facilitate protein deposition. Primary human bone marrow stromal cells (hBMSCs, from 24 year old Female, Tulane Center for Gene Therapy) were cultured at 37°C with 5% by volume CO2 in culture medium. For cell seeding, hBMSCs were allowed to achieve 80% confluence in a T-75 culture flask before dissociation with 0.25% mass fraction trypsin (containing 1mmol/L ethylenediaminetetraactate (EDTA)) and re-suspended in culture medium. Cell number was determined using a hemocytometer. Passage 5 cells were seeded onto scaffolds in 48-well plates at 10,000cells/cm2 (scaffolds for cell imaging and alkaline phosphatase activity assay were seeded at 5,000cells/cm2). Cells were seeded in 500µL of cell suspension in culture medium per well. Positive control TCPS+osteogenic supplement (OS) cultures were supplemented with 10nmol/L dexamethasone, 20mmol/L β-glycerophosphate and 0.05mmol/L ascorbic acid. Medium was changed twice per week with culture medium or culture medium supplemented with OS. Cells were cultured at 1 d, 4 d, 14 d, and 50 d as indicated.
Picogreen DNA assay
The Picogreen DNA assay was used to assess cell proliferation by measuring DNA concentration. At 1d and 14d time points, samples (including no cell controls) were rinsed with PBS then frozen overnight at -20°C to rupture cell membranes. Cultures were then digested for 16h at 60°C in digestion buffer (0.1mL papain solution (activity 35U/mg) and 35mg cysteine in 20mL PBS). Cell lysate solution was then transferred to a 96-well plate and diluted 1:1 in Picogreen reagent (Invitrogen, 200µL). DNA fluorescence (excitation 485nm, emission 515nm) was measured using a plate reader and calibrated using a DNA standard curve.
Alkaline phosphatase assay
Alkaline phosphatase (ALP) concentration was detected using the Stanbio alkaline phosphatase assay kit according to kit procedures. ALP was measured at 4 and 14d time points. Briefly, samples and controls (TCPS and TCPS+OS) were rinsed with PBS (37°C). Cells were then lysed by incubating in 10mL/L NP-40 detergent (Stanbio) in DI water for 10 min at 37°C. 50µL of cell lysate was assayed using the p-Nitrophenylphosphate (17.0mmol/L) reaction solution in 24-well plates. NP-40 solution with reagent was used as a blank and an ALP standard curve was generated from freshly reconstituted control sera with known alkaline phosphatase levels (Normal Control Serum, StanbioSer-T-Fy 1). Absorbance values were read on a plate reader at 405nm every 20min for 5h. Change in normalized absorbance per minute was calculated from a linear region of the curve and used to calculate units per liter of ALP. ALP concentration was then normalized to DNA concentration from parallel cultures (Picogreen DNA assay).
Alizarin red (AR) staining for mineral deposition
Cultures were fixed in 37g/L formaldehyde in phosphate buffered saline (PBS, 37°C) for 20 min, rinsed with DI water then stained (1h) for mineral deposition with Alizarin red solution (0.5mg/mL in DIwater). Samples were rinsed extensively with DI water to remove excess stain (rinse was considered complete when paired control samples with no cells did not show distinct staining patterns). Digital images (5X magnification) of stained samples were acquired using a stereomicroscope. Four samples from each treatment group were imaged for their mineral deposition.
Confocal fluorescence imaging of cells and cell shape analysis
Confocal microscopy (Leica SP5) was used to characterize cell morphology of hBMSCs cultured for 1d on NF, MOD NF, SC and MOD SC samples (cell seeding density, 5,000cells/cm2). Cells were fixed in 37 g/L formaldehyde in phosphate buffered saline (PBS, 37°C) for 1 h, rinsed with fresh PBS, then permeablized with 1g/L Triton X-100 solution for 10min. Samples were then rinsed with blocking buffer (50g/L bovine serum albumin (BSA)) for 30min. Actin cytoskeleton was fluorescently labeled with Alexa Fluor 546-phalloidin (0.33µmol/L in blocking buffer, incubated for 30min) while the nucleus was stained with DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) fluorescent stain (0.03mmol/L in 10g/L BSA, incubated for 5min). Samples were rinsed in PBS before imaging. High resolution z-stack images were captured with a 63x/0.90 numerical aperture (NA) water immersion objective (1024 x 1024 resolution, 1µm z-step size). High-resolution confocal z-stack images of cells were necessary in order to capture fine feature of the cell boundary and also to capture cell processes that penetrate the porous NF scaffold. Only cells with no cell-cell contacts were analyzed to clearly delineate individual cell shape. Three samples were prepared for each treatment and over 35 cells were imaged and analyzed for each sample type.
Cell outlines were obtained by implementing a modified snake algorithm27 as previously described by Driscoll et al.28,29 (Supplemental Information M2) A total of 10 shape descriptors were calculated including cell area (µm2), perimeter (µm), mean boundary distance (µm), major axis length (µm), mean negative curvature (1/µm), regions of negative curvature (normalized to perimeter), tortuosity, circularity, solidity, aspect ratio. For a description of cell shape metrics, (Supplementary Table S1).
Human genome microarray
Total RNA from hBMSCs cultures were collected at 14d (Supplemental Information M3). Whole genome transcriptions of hBMSCs were measured by high-throughput microarray screening using the Illumina Human HT-12 v4 Expression Bead Chips (IL1-HTH-110) with 47,231 probes through a contract with Expression Analysis Inc. (Durham, NC). Prior to microarray screening NIST ERCC spike-in controls were added to samples to monitor the quality of microarray data acquisition.
Microarray data was analyzed using the BRB-Array Tools developed by Dr. Richard Simon and the BRB Array Tools Development Team (version 4.1.0, Biometric Research Branch, National Cancer Institute) (Supplemental Information M4). From a total of 47,231 genes identified with the microarray, a subset of 199 genes was found to exhibit at least a 1.5-fold change from the median value of each gene (Figure 7A). The “significance analysis of microarrays” (SAM)30 method was used to identify differentially expressed genes across class comparisons. Scaffolds were grouped into classes of ALL NF (NF and MOD NF), ALL SC (SC and MOD SC), ALL MOD (MOD NF and MOD SC), and ALL UN-MOD (NF and SC) for investigation of the effects of structure and chemistry on gene expression. Differentially expressed genes from SAM analysis were used for further gene ontology-based (GO) annotation in the Database for Annotation, Visualization, and Integrated Discovery (DAVID)31,32 (Supplemental Information M2). Average fold change values for select individual genes were calculated based on their log2 fold change from the average TCPS control value for that gene (Supplemental Information M5).
Chemistry of scaffolds modified via mild hydrolysis
Water contact angle (WCA) dropped significantly with mild hydrolysis treatment on both NF scaffolds and SC films (from 145°±6°to 112°±4° on NF to MOD NF and 75°±7° to 48°±7°on SC to MOD SC) (Figure 2A). It should be noted however that contact angle measurements on nanofiber surfaces cannot provide a quantitative measure of surface energy due to the porous and irregular nature of the fiber mat surface.35 A corresponding increase in COOH concentration was also detected on NF scaffolds where COOH concentration increased from 1.6µmol/L±1.1µmol/L on NF scaffolds to 6.6 µmol/L±1.1µmol/L on MOD NF scaffolds (Figure 2A). No significant increase in COOH concentration was detected on MOD SC films compared to SC film. Elemental survey scans of the scaffold surfaces via XPS detected an increase in O 1s on MOD NF scaffolds compared to NF scaffolds due to opening of carboxyl groups and hydroxyl groups on the MOD NF scaffold surface (Figure 2B).19 In addition, Na 1s was detected on MOD NF scaffolds and MOD SC films. The overall O 1s to C 1s ratio was increased significantly on MOD NF scaffolds compared to NF scaffolds (Figure 2C). There was no significantly detected increase on O 1s species on MOD SC films compared to SC films. It can be assumed however, based on the mechanisms of base hydrolysis as well as the significant decrease in water contact angle on modified SC films, that carboxyl and hydroxyl groups were generated at physically significant concentrations on the SC film material surface. For further analysis of these results, (Supplemental Information R1).
Structure of NaOH modified scaffolds

SEM images demonstrate the similarities between overall NF scaffold structure between NF scaffolds and MOD NF scaffolds, and maintenance of individual nanofiber morphology (Figure 3A). In MOD NF scaffolds, there is no evidence of fiber pitting or breakage typically seen with alkaline hydrolysis of PCL (Supplementary Figure S1). Mean fiber Diameter and fiber diameter variability (represented by the coefficient of variation, Cν) were quantified from SEM images (magnification 5000) (Figure 3B). Average nanofiber diameter for NF and MOD NF scaffolds was 611µm and 600µm respectively with Cν of 0.142 and 0.169 respectively. Mean fiber diameter and variance in fiber diameter between NF and MOD NF scaffolds were not found to be statistically different (Figure 3B). SC Films were also chemically modified by NaOH hydrolysis to alter surface chemistry while maintaining the overall film structure. SC films were imaged at magnification 1000 and magnification 50,000 to monitor the film structure with NaOH hydrolysis (Figure 3A). Films maintained their characteristic cobble stone pattern seen before and after chemical modification.

Effect of scaffold structure and chemistry on hBMSC proliferation
No significant differences in DNA concentration were seen after one day of culture (Figure 4). After 14 d of culture DNA concentration increased as compared to Day 1 for all scaffolds, indicating that hBMSCs attached and proliferated on all scaffolds investigated. At 14 d of culture, there were no significant differences between DNA concentration on NF versus MOD NF or SC versus MOD SC, indicating that presence of -COOH/-OH groups on the scaffold surface had no effect on hBMSC proliferation (Figure 4). There was however a trend of increased cell proliferation on SC films compared to NF scaffolds, with MOD SC films having significantly higher DNA concentration than MOD NF films (Figure 4). Keselowsky et al.39 saw similar results when investigating osteoblast-like cells (MC3T3-E1) on self-assembled monolayers of -OH, -COOH, -CH3, and -NH chemistries, where no significant differences in cell proliferation were observed with varying surface chemistry.39 This is a particularly important outcome, as it allows the comparison of other biological outcomes which may be influenced by cell viability and proliferation (i.e. mineralization and differentiation assays).

Differentiation of hBMSCs on chemically modified scaffolds
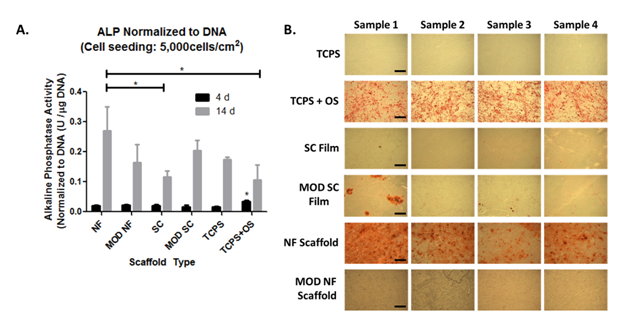
Alkaline phosphatase (ALP) is a metalloenzyme implicated in the early stages of mineralization and often used as a biomarker for osteogenic differentiation.40,41 ALP activity was significantly higher in TCPS+OS controls at 4d compared to all other substrates (Figure 5A). At 14d ALP activity on NF scaffolds was significantly higher than on SC films indicating higher osteogenic potential on the NF scaffolds. ALP activity in TCPS+OS samples was significantly less than on NF samples at 14d (Figure 5A). This result is not surprising as ALP expression is an early marker of osteogenic differentiation and is known to decrease as mineralization progresses.42 MOD NF scaffolds did not have significantly higher ALP expression than SC or MOD SC films at 14 d (Figure 5A). These trends were further supported by 50d alizarin red staining for calcified mineral.
After 50d of culture, NF scaffolds showed strong alizarin red staining even without osteogenic supplement, demonstrating the ability of these scaffolds to support osteogenic differentiation (Figure 5B). MOD NF scaffolds however, did not show appreciable alizarin red staining, in accordance with their lower ALP expression at 14 d (Figure 5). In a complimentary study (Supplementary Figure S2) we found that MOD NF scaffolds could support hBMSC mineralization in the presence of osteogenic supplement. Both SC films and MOD SC films showed little to no signs of calcified mineral after 50 d of culture, similar to previously reported studies.14 Positive TCPS+OS controls demonstrated a strong presence of calcified mineral, while negative TCPS controls did not have observable mineral deposition (Figure 5B).
Confocal fluorescence imaging of hBMSCs
Cell shape was investigated after 24 h of culture for cells on NF, MOD NF, SC and MOD SC scaffolds (Figure 6). 10 shape metrics were determined from snake outlines of the maximum projections of 63x/0.9 NA confocal z-stack images (n>35 cells per scaffold type).Significant differences between all possible pair wise comparisons were determined using a 1-Way ANOVA with Tukey’s post-test (Table 1) and (Supplementary Figure S3) for cell shape metric values. All cell shape metrics for NF vs SC scaffolds were significantly different (p<0.05) except for aspect ratio (p>0.05), which was significantly different between NF and MOD SC (p<0.05). For NF vs. MOD NF, fewer significantly different metrics were identified, including regions of negative curvature (p<0.001), tortuosity (p<0.01), solidity (p<0.01) and aspect ratio (p<0.05). The effect of scaffold chemistry on cell shape was more pronounced in the NF system compared to the SC system. For SC vs. MOD SC, there were no significant differences in cell shape metrics (p>0.05) (Table 1). These results indicate a stronger dependence of cell shape on scaffold structure than on scaffold chemistry.
|
Area (µm2) |
Perimeter(µm) |
Mean boundary distance(µm) |
Mean negative curvature |
Regions of negative curvature |
Tortuosity |
Circularity |
Solidity |
Aspect ratio |
Minor axis length(µm) |
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
(µm-1 ) |
|
|
|
|
|
|
NF vs.MOD NF |
ns |
ns |
ns |
ns |
*** |
** |
ns |
** |
* |
ns |
NF vs.SC |
*** |
** |
*** |
** |
* |
** |
*** |
*** |
ns |
*** |
NF vs. MOD SC |
*** |
ns |
** |
*** |
ns |
*** |
*** |
*** |
* |
*** |
MOD NF vs. SC |
*** |
** |
* |
** |
ns |
** |
** |
ns |
*** |
*** |
MOD NF vs. MOD SC |
*** |
ns |
ns |
*** |
ns |
** |
*** |
** |
*** |
*** |
SC vs. MOD SC |
ns |
ns |
ns |
ns |
ns |
ns |
ns |
ns |
ns |
ns |
Table 1 Summary of significant difference in cell shape metrics. Shape metrics were determined from snake outlines of hBMSCs cultured on scaffolds of varying chemistry and structure. Statistical differences across four scaffold groups were analyzed using 1-way ANOVA with Tukey’s multiple comparisons post-test (ns p>0.05, * p<0.05, ** p<0.01, *** p<0.001, n>35)*
NF, PCL non-woven nanofibers; SC films, PCL spin-coated TCPS discs; MOD NF, NF scaffolds modified by mild hydrolysis; MOD SC, SC films modified by mild hydrolysis.

Human genome microarray
Microarray analysis of mRNA expression after 14d of culture found a subset of 199 genes to be significantly expressed (passing a 1.5 fold, 10% filter) across all samples, relative to an average TCPS reference array (Figure 7A). Hierarchical clustering occurred within replicates with limited correlations across sample type. Through DAVID annotation cluster analysis, Biological process GO Terms relating to osteogenic phenotype were identified with a significant enrichment score of 2.6 (Figure 7B). Further, the gene for alkaline phosphatase (ALPL) was present in this annotation cluster. ALPL log2 fold change from TCPS (Figure 7C) was found to follow a similar trend to that seen in biochemical analysis of alkaline phosphatase activity at 14d (Figure 5A). ALPL gene fold change was significantly higher on NF than that of MOD NF, MOD SC, and TCPS+OS.TCPS+OS ALPL fold change was negative indicating a decrease in ALPL expression at 14d with osteogenic supplement. This result is in agreement with a decreased ALP activity for TCPS+OS seen with biochemical analysis at 14 d (Figure 5A).

Scaffolds were grouped into classes of ALL NF (NF and MOD NF), ALL SC (SC and MOD SC), ALL MOD (MOD NF and MOD SC), and ALL UN-MOD (NF and SC) for investigation of the effects of structure and chemistry on gene expression. SAM class comparisons were conducted on the 199 significantly expressed genes comparing ALL NF vs ALL SC and ALL MOD vs ALL UN-MOD. SAM analysis resulted in 16 differentially expressed genes between ALL NF and ALL SC, and 74 differentially expressed genes between ALL MOD vs ALL UN-MOD.
Annotation cluster analysis of GO term enriched from ALL NF vs ALL SC genes resulted in one significant annotation cluster (enrichment score of 2.0). The annotation cluster contained GO terms related to extracellular regions, parts, and matrix as well as proteinaceous extracellular matrix (Table 2). Genes found in this annotation cluster are presented in Figure 8A as log2 fold change from TCPS. ECM related genes, COL8A2, FNDC1, FBLN5, and GPC4, were all found to be down regulated on SC films, regardless of chemical modification, while these genes were up regulated on NF scaffolds regardless of chemical modification (Figure 8A). IGFBP1 which promotes cell migration and alters the interaction of IGFs with cell surface receptors.43 was up regulated in SC and MOD SC films while it was minimally regulated in NF and MOD NF scaffolds. IL 8, generally released in response to inflammatory stimulus,44 was largely up regulated in SC, MOD SC, and NF scaffolds while it was not regulated in MOD NF scaffolds (Figure 8A).
Effect of structure: all NF vs. all sc ( regardless of chemistry) |
||
|---|---|---|
GOTERM: cellular components |
||
Annotations cluster 1 enrichment score 2.0 |
Gene count |
P-value |
extracellular region part |
5 |
0.0017 |
extracellular region |
6 |
0.0035 |
proteinaceous extracellular matrix |
3 |
0.016 |
extracellular matrix |
3 |
0.018 |
Effect of chemistry: all MOD vs. all UN-MOD(regardless of structure) |
||
KEGG pathway |
Gene count |
p-Value |
Glycolysis/ Gluconeogensis |
9 |
1.5×10-9 |
Fructose and mannose metabolism |
7 |
3.7× 10-8 |
Pentose phosphate pathway |
4 |
4.3×10-4 |
GOTERM: Biological process |
||
Annotation cluster 1 enrishment score: 7.6 |
Gene count |
P-value |
Glycolysis |
8 |
2.9×10-10 |
monosaccharide metabolic process |
12 |
3.1×10-10 |
hexose metabolic process |
11 |
1.3×10-9 |
Annotation cluster 2 enrishment score: 2.8 |
Gene count |
P-value |
Monosaccharide biosynthesis process |
4 |
3.2×10-4 |
pyruvate metabolic process |
4 |
4.6×10-4 |
alcohol biosynthesis process |
4 |
5.3×10-4 |
Annotation cluster 3 enrishment score: 2.1 |
Gene count |
P-value |
carboxylic acid biosynthesis process |
5 |
0.0024 |
organic acid biosynthesis process |
5 |
0.0024 |
amine biosynthesis process |
4 |
0.0031 |
Gene count |
P-value |
|
Annotation cluster 4 enrishment score: 1.3 |
||
protein oligomerization |
4 |
0.0025 |
protein complex biogenesis |
6 |
0.0035 |
protein complex assembly |
6 |
0.0035 |
Annotation cluster 5 enrishment score: 1.3 |
Gene count |
P-value |
negative regulation of cell migration |
3 |
0.018 |
negative regulation of locomotion |
3 |
0.021 |
negative regulation of cell motion |
3 |
0.022 |
Table 2 Functional Annotation Clustering using DAVID analysis of genes differentially expressed between ALL NF vs ALL SC (16 genes found) and between ALL MOD vs ALL UN-MOD (79 genes found). “ALL NF” combines gene expression data from NF and MOD NF, “ALL SC” combines SC and MOD SC, “All MOD” combines MOD NF and MOD SC, and “ALL UN-MOD” combines NF and SC*.
NF, PCL non-woven nanofibers, SC films, PCL spin-coated tcps discs; MOD NF, NF scaffolds modified by mild hydrolysis; MOD SC, SC Films modified by mild hydrolysis; MOD SC, SC films modified by mild hydrolysis; ALL NF, NF and MOD NF; ALL SC, SC and MOD SC; ALL MOD, MOD NF and MOD SC; ALL UN-MOD, NF and SC; KEGG, kyoto encyclopedia of genes and genomes; GOTERM, gene ontology term

Annotation cluster analysis of KEGG pathways enriched from ALL MOD vs ALL UN-MOD genes resulted in pathways primarily related to glycolysis/gluconeogenesis and sugar metabolism (Table 2). Also enriched was a KEGG pathway related to pathogenic Escherichia coli (E. coli) infection, however, the specific genes enriched from SAM analysis are also related to actin cytoskeletal reorganization. Biological process GO term cluster analysis also similarly resulted in significantly enriched clusters related to glycolysis, sugar processing and cell migration/locomotion (Table 2).The specific genes found in the enriched KEGG pathways are presented in Figure 8B & C. KEGG pathway genes related to glycolysis and sugar processing were found to be generally up-regulated or minimally-regulated in NF and SC scaffolds, however they are largely down-regulated in MOD NF and MOD SC scaffolds (Figure 8B).
Several genes related to cytoskeletal organization were also significantly enriched into KEGG pathways: KRT18 (keratin 18), RHOA (ras homolog gene family, member A), TUBB6 (tubulin, beta 6), and TUBBP1 (similar to tubulin, beta 5). These genes were identified as a part of the pathogenic Escherichia coli infection KEGG pathway, however, these genes also strongly associate with the cytoskeleton gene ontology for cellular components (Gene count 4, p-value of 0.0013). There were no infection related genes identified as a part of the E.coli pathway, suggesting that the regulation of these cytoskeletal genes was not a result of infection. Cytoskeleton related genes were consistently up-regulated in un-modified scaffolds while they were minimally-regulated or down-regulated in modified scaffolds (Figure 8C).
Nanofiber scaffolds have shown particular promise in bone tissue engineering and regenerative medicine strategies. Enhanced differentiation of stem cells in nanofiber scaffolds has been previously demonstrated, however the material properties and cellular mechanisms driving this process are largely unknown.7,9-13 In this study, we have comprehensively investigated hBMSC response to nanofiber scaffolds of different chemistry to examine the contributions of scaffold structure and chemistry on stem cell response.
To minimize differences in nanofiber scaffold structure while varying nanofiber chemistry, we employed a wet chemical method to modify the nanofiber mat surface after the formation of fibers. After mild hydrolysis with NaOH treatment, nanofiber scaffolds demonstrated decreased water contact angle with concurrent increase in –COOH functional groups on the scaffold surface while SEM images indicated there were no significant changes to fiber diameter or overall nanofiber scaffold structure. PCL spun coat films were also investigated to serve as 2-D substrate analogs of the nanofiber scaffolds. Overall, a water contact angle decrease of approximately 33° was seen on MOD NF scaffolds while a decrease of approximately 27˚ was detected on MOD SC films. It has been previously reported that changes in water contact angle of this magnitude on 2-D surfaces can produce relevant changes in cell behaviors such as cell shape, attachment, proliferation and differentiation.45,46
Modification of the nanofiber scaffold surface chemistry resulted in significant changes to hBMSC osteogenic differentiation. hBMSCs on MOD NF scaffolds did not show the characteristic osteogenic response (elevated alkaline phosphatase activity and mineralization) generally observed on NF scaffolds (Figure 5).14 A decrease in osteogenic response with the introduction of surface -COOH/-OH functional groups on the nanofiber scaffolds is consistent with reports for 2-D -COOH functionalized materials. In a study by Keselowsky et al.39 self-assembled monolayers of varying functionality were coated with fibronectin then investigated for their effect on the expression of osteoblast specific genes by MC3T3-E1 cells. They found that gene markers for ALP, bone sialoprotein and osteocalcin were all down regulated on -COOH surfaces.39 Similarly, Curran et al.44 found that 2-D –OH and -COOH surfaces did not support osteogenesis of bone-marrow-derived mesenchymal stem cell while they did promote chondrogenesis.44 These results suggest that the nanofiber scaffold structural cues cannot overcome surface chemistry cues for cell response.
The current material system provides a unique opportunity to investigate the mechanisms behind osteogenic differentiation in nanofiber scaffolds. One possible mechanism is the influence of scaffold architecture on cell shape.14,46 Several studies have demonstrated a possible correlation between cell shape and stem cell differentiation .47–50 In nanofiber scaffolds particularly, Kumar et al.14 noted that 1 day hBMSC shape was similar to that on flat substrates treated with osteogenic supplements and suggested that the osteo inductive effects of nanofibers come from their ability to elicit an osteogenic morphology.14 In our system, we were able to investigate cell shapes in nanofiber scaffolds that are associated with both osteogenic, and non-osteogenic outcomes. In the NF scaffold, we observe positive expression of markers associated with osteogenic differentiation, while in the MOD NF scaffold markers of osteogenic differentiation are reduced or show limited expression. Interestingly, in investigating 1 d cell shape between the two materials, we find that only a few metrics are significantly different, while the majority of metrics are unaltered by the chemical modification and subsequent state of hBMSC differentiation.
Metrics found to be significantly different between NF and MOD NF includes regions of negative curvature, tortuosity, solidity, and to a lesser extent aspect ratio (Table 1). These metrics which are significantly different between cells in an osteogenic nanofiber environment and a non-osteogenic nanofiber environment may be possible targets for the use of cell shape metrics in the prediction of osteogenic differentiation in nanofiber scaffolds. On the other hand, metrics such as cell projected area, perimeter and circularity have been demonstrated here to not be strongly correlated with osteogenic differentiation in the nanofiber system. These results differ from that seen in flat 2-D culture environments, where the analogous metrics of cell spread area and roundness were found to be strongly correlated to stem cell lineage commitment.48 Therefore, specific cell metrics relevant to differentiation in 2-D may not directly translate to a more 3-D topographical system where the scaffold architecture is dominant in determining cell shape.
In our material system we can also investigate the relative contributions of structure and chemistry in directing stem cell response. Through microarray gene expression analysis, we find that ECM related pathways are more affected by scaffold structure while metabolic and cytoskeletal related pathways are more affected by scaffold surface chemistry. Although gene expression values from microarray data are only semi-quantitative, genes identified in these pathways provide candidates for future investigation (through PCR and immune staining) of specific cellular mechanisms that may be associated with hBMSC response to nanofiber scaffold properties.
ECM related genes (COL8A2, FNDC1, FBLN5 and GPC4) were generally up-regulated on all NF scaffolds (NF and MOD NF) regardless of scaffold surface chemistry (Figure 8A). In contrast, the same genes are consistently down-regulated on spun coat films (SC and MOD SC). Interestingly, the up-regulation of ECM related genes has been demonstrated to be correlated with in vivo bone forming phenotype of hBMSCs.51 Lui et al.52 also investigated microarray gene expression analysis of hBMSCs in nanofiber scaffolds and found ECM GO Terms to be significantly enriched on nanofiber mats.52 Surprisingly, in our study, the regulation of ECM related genes was independent of the osteogenic outcome (Figure 5B). These results suggest that scaffold structure (NF versus SC Film) is a dominant scaffold feature in ECM related gene regulation, but not necessarily in vitro osteogenic response.
Scaffold surface chemistry was found to influence gene expression in pathways related to glycolysis/gluconeogenesis and other metabolic pathways (Table 2). Scaffold chemical modification (mild hydrolysis inducing –COOH/-OH groups) resulted in down-regulation of genes associated with glycolysis/gluconeogenesis and metabolism regardless of scaffold structure (Figure 8B) whereas, only un-modified scaffolds showed up-regulation of several of these genes (PFKFB4, TPI1, ENO2, PCK2, and PFKP). It should be noted that the glycolysis/gluconeogenesis pathway shares many overlapping genes with other pathways, making interpretation of the functional outcome of these results difficult. For example, the adolase family of genes is also associated with gene ontologies related to actin cystoskeleton and cytoskeletal binding and have been implicated in the regulation of cell contraction.53 Scaffold chemistry resulted in regulation of these related genes independent of both scaffold structure and osteogenic response (Figure 8B).
Surface chemistry and hydrophilicity affect both the amount of adsorbed serum proteins as well as the conformation of proteins at the cell-biomaterial interface.54–57 The increase of surface carboxyl groups in particular has been associated with increased protein adsorption from serum containing culture media as well as conformational changes in the adsorption of specific proteins such as fibronectin.54–57 It is possible that the presence of carboxyl and hydroxyl groups on the modified nanofiber and spun coat film surfaces may have altered protein adsorption to the scaffold, thereby altering the presentation of key proteins, cytokines and growth factors related to signaling in the metabolic pathways.
In addition to the enrichment of metabolic pathways, expression of genes related to the cytoskeleton was also influenced by surface chemistry. KRT18, RHOA, TUBB6, and TUBBP1 were generally found to be up regulated in NF and SC scaffolds while these genes had relatively low expression in the MOD NF and MOD SC scaffolds (Figure 8C) indicating an influence on cytoskeletal organization by –COOH/-OH chemical modification. Cytoskeletal regulation is consistent with our finding that cell shape is moderately affected by nanofiber chemical modification. Although cell shape was more strongly affected by scaffold structure (NF vs SC) (Table 1), genomic regulation of the cytoskeleton in UN-MOD vs MOD scaffolds indicates changes in the cytoskeleton as a possible mechanism through which scaffold chemistry may alter stem cell function. In particular, cell shape was demonstrated by Mc Beath et al.48 to play an integral role in stem cell linage commitment through the modulation of Rho A activity as well as the generation of cytoskeletal tension.48 In our study, we find that RHOA is up-regulated in the un-modified scaffolds (NF and SC) while it is minimally expressed or even down- regulated (MOD SC and MOD NF) in the modified scaffolds relative to TCPS controls (Figure 8C). The Rho A pathway as well as the generation or disruption of cytoskeletal tension should be further investigated as a possible mechanism underlying scaffold chemistry-related modulation of stem cell lineage commitment.
We have developed a material system that allows investigation of the effects of both scaffold structure and chemistry in regulating stem cell response in the nanofiber scaffold environment. The nanofiber environment can promote osteogenic differentiation; however, alterations in scaffold chemistry can modulate this response. Nanofiber surface chemical modification with COOH/OH via mild hydrolysis results in reduced osteogenic response from hBSMCs. These studies suggest that both nanofiber scaffold structure and chemistry should be carefully engineered in order to achieve a desired biological response. Cell response to modified nanofiber scaffolds are reflected in changes to cell shape, cytoskeletal rearrangement and metabolic changes. Nanofiber scaffold structure was found to influence cell shape and ECM gene expression; however, these cellular responses may not be directly correlated to osteogenic response. The MOD NF scaffolds provide a negative control for osteogenic differentiation in the NF scaffold system, thereby allowing for further investigation into the mechanisms behind nanofiber scaffold mediated osteogenic differentiation.
None.
S. Sarkar was supported by the National Research Council-NIST postdoctoral fellowship. B.A. Baker was supported by the National Research Council-ARRA-NIST postdoctoral fellowship. Thank you to Christopher M. Stafford, NIST, for assistance with XPS.
Official contribution of the National Institute of Standards and Technology; not subject to copyright in the United States.

©2016 Sarkar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.