Advances in
eISSN: 2373-6402


Review Article Volume 8 Issue 1
Faculty of Agriculture and Food Sciences, Universiti Putra Malaysia, Malaysia
Correspondence: Patricia King Jie Hung, Faculty of Agriculture and Food Sciences, University Putra Malaysia, Bintulu, Sarawak Campus, P.O Box 396, Nyabau Road,97008, Malaysia
Received: October 10, 2017 | Published: February 14, 2018
Citation: Khai CTZ, Ming SC, Hung PK. They are different: molecular approach on Tirathaba pest infesting oil palm and coconut tree. Adv Plants Agric Res. 2018;8(1):71-73. DOI: 10.15406/apar.2018.08.00294
There are some confusion among agriculturists on the species of Tirathba beetles that are infesting on oil palm and coconut trees. Many thought they are the same species. In this study, the mitochondrial DNA Cytochrome oxidase subunit I (COI) of Tirathaba pest infested oil palm and coconut tree were compared. The mitochondrial DNA Cytochrome oxidase subunit I (COI) gene of the targeted Tirathaba sp. infesting on oil palm and coconut tree were sequenced. The sequences were trimmed to remove gaps and produce a final aligned fragment of 603bp for oil palm Tirathaba sample and 602bp for coconut pest sample.The DNA sequences were analyzed with other Tirathaba sp.sequences available in Gene bank using phylogenetic tree constructed with Neighbor-Joining (NJ) and genetic distance analysis algorithms. The result of this study indicates they were two different species. This knowledge will provide important data elements in the development of pest management strategy.
High infestation rate of Tirathaba mundella was reported from many oil palm plantations established on peat Masijan et al.1 The infestation is identified by the presence of long tubes of silk and frass which is built by the pest. The larvae stage of pest does most of the damage by feeding or scraping leaving holes on immature fruitlets. Larvae of early instar stage often attack on male inflorescences while larvae of older instar stage attack on female inflorescences and bunch Yaakop et al.2 A new damage caused by Tirathaba mundella is visible as moist and reddish brown faeces on brunches while an old damage is characterized as dry and brownish black faeces. Another Tirathaba that also regarded as agricultural pest is Tirathaba fructivora. It was first observed in Fiji and Java, Indonesia Paine3 and later in Sumatra, Indonesia and Malaysia (Kalshoven, I981). The pest causes destructive effect to the coconut plantation in Philippines. Tirathaba fructivora together with another coconut spike moth, Tirathaba rufivena Walker are making damage to coconut flower. Both pests attack the inflorescence of coconut, causing the young and soft flower to drop off from the plant. Falling of flower prevents the formation of nuts Alouw et al.4 Tirathaba fructivora has a total development period of around 25days from the stage of egg deposition to adult emergence. The newly laid egg of Tirathaba fructivora is white in colour and it turns dark yellow or dark orange when it is about to hatch. Hatching took about 4days from egg deposition. The larvae have five instars level with a larval stadium of around I7days and pupal period of around 8days.
The first instar larvae are light brown and it turns into darker colour slowly in the following instars. Thoracic shield was developed on the first thoracic segment from first instars and becomes more apparent in the later instars. Prolegs are well grown from the second to fifth instars. The longevity of female and male Tirathaba fructivora is around 8-9days Alouw et al.4 Many authors believe that the Tirathaba pest found in both plantation were the same species. This notion needs to be clarified. Therefore this study aims to compare the genetic markers CO1 of both Tirathaba sp. infesting on the oil palm and coconut tree. The finding of this study provides essential data to determine if the Tirathaba species from both type of plantation belongs to same species.
Tirathaba mundella were collected in the Sarawak Oil Palms Berhad (SOP) plantation site, located in Miri, Sarawak. The specimens were collected from the male flower. For the coconut pest, the insect were collected from immature nuts of coconut tree. Tirathaba sp. from both plantations were DNA extracted, amplified with primers coding for the cytochrome oxidase I (COI) gene, which were “LepFI” 5’-ATTCAACCAATCATAAAGATATTGG-3’ and “LepRI” 5’-TAAACTTCTGGATGTCCAAAAAATCA-3’ (Hajibabaei et al. 2006). The amplified DNA was sequenced and blasted against public available Gene bank. The sequences were further analyzed their genetic distance with other Tirathaba spp. using the Kimura two parameter Hosoishi et al.5 Kimura.6 A phylogeny tree was constructed by using Neighbor-Joining (NJ) method Saitou et al.,7 in the program MEGA version 7 Hosoishi et al.5
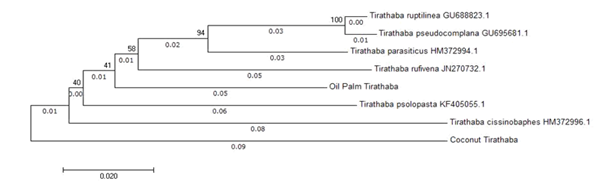
The sequenced DNA fragments for both samples collected from oil palm and coconuts were approximately 602 base pairs (bp). The BLAST result showed only Tirathaba sp from oil palm plantation had only 92% of similarities with Tirathaba parasiticus sequence in the Gene bank while coconut pest sample showed less than 95% similarities with other Tirathaba spp available in the database (similarities>95%). The genetic distances for both Tirathaba spp were analyzed with other six Tirathaba sp. The six species included Tirathaba pseudocomplana, Tirathaba rufivena, Tirathaba parasiticus, Tirathaba ruptilinea, Tirathaba cissinobaphes and Tirathaba psolopasta. All Tirathaba sp. COI sequences that contained more than 600bp were aligned by Multiple Sequence Alignment Tool (MUSCLE) and result as shown in Table 1. A phylogeny tree was generated (Figure 1) and shows that the oil palm Tirathaba sample was under the same clade with Tirathaba pseudocomplana, Tirathaba rufivena, Tirathaba parasiticus and Tirathaba ruptilinea.
S. No |
Genetic diversity of Cytochrome c Oxidase subunit i sequence |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
1 |
Tirathaba ruptilinea |
|||||||
GU688823.I |
||||||||
2 |
Tirathaba rufivena |
0.I0I5 |
||||||
JN270732.I |
||||||||
3 |
Tirathaba psolopasta |
0.II29 |
0.II33 |
|||||
KF405055.I |
||||||||
4 |
Tirathaba pseudocomplana |
0.0II8 |
0.0976 |
0.II49 |
||||
GU69568I.I |
||||||||
5 |
Tirathaba parasiticus |
0.0644 |
0.I0I5 |
0.I229 |
0.07 |
|||
HM372994.I |
||||||||
6 |
Tirathaba cissinobaphes |
0.I449 |
0.I470 |
0.I486 |
0.I5I2 |
0.I367 |
||
HM372996.I |
||||||||
7 |
Oil Palm Tirathaba mundella |
0.II3I |
0.I036 |
0.I2I3 |
0.IIII |
0.0899 |
0.I424 |
|
8 |
Coconut Tirathaba |
0.I732 |
0.I820 |
0.I628 |
0.I775 |
0.I7I2 |
0.I840 |
0.I485 |
Table 1 Estimates of evolutionary divergence between sequences
Tirathaba mundella showed a genetic distance with the coconut Tirathaba at 0.I485. This indicates that the two species were not from the same genus. The Blast result also indicates that the pest infested the coconut plantation could be misidentified as Tirathaba rufivena as their COI sequence were distantly related. The coconut pest may be a newly discovered Tirathaba species which needed to further study.8
None.
The author declares no conflict of interest.

©2018 Khai, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.