Advances in
eISSN: 2373-6402


Research Article Volume 9 Issue 1
1Hybrid Rice Division, Bangladesh Rice Research Institute, Bangladesh
2Genetic Resources and Seed Division, Bangladesh Rice Research Institute, Bangladesh
3Agricultural Statistics Division, Bangladesh Rice Research Institute, Bangladesh
4Department of Entomology, Sher-e-Bangla Agricultural University, Bangladesh
5Agronomy Division, Bangladesh Rice Research Institute (BRRI), Bangladesh
Correspondence: Anowara Akter, Hybrid Rice Division, Bangladesh Rice Research Institute, Gazipur- 1701, Bangladesh
Received: January 30, 2018 | Published: January 4, 2019
Citation: Akter A, Hasan MJ, Kulsum MU, et al. Stability and adaptability of promising hybrid rice genotypes in different locations of Bangladesh. Adv Plants Agric Res. 2019;9(1):35-39. DOI: 10.15406/apar.2019.09.00407
Over environments, identifying superior genotypes and most discriminating environments and thus Assessing the stability performance and genotype x environment interaction is one of the important steps for accurate rice promising genotypes evaluation in large multi-environment trials. In this study, six rice promising genotypes were grown in four environments during 2009 plant season to determine the grain yield stability and adaptability. The experiment used randomized complete block design with three replications. Yield stability and adaptability of yield performance were analyzed by combined analysis and AMMI model. From the current study the combined analysis of variance for grain yield revealed highly significant (P<0.01) for genotypes, environments and their interactions. The significant interaction indicated that the genotypes respond differently across the different environments. The mean grain yield of genotypes over environments indicated that BRRI 10A/ BRRI 11R (G3) and BRRI hybrid dhan2 (G4) had the highest yield (7.43 tha-1) and (7.84 tha-1) while genotype, BRRI dhan28 (G5) had the lowest yield (5.14 tha-1) respectively. In AMMI analysis, AMMI 1 biplot showed the hybrids BRRI 10A/BRRI11R (G3) and BRRI hybrid dhan2 (G4) had higher mean yields with high main (additive) effects. Hence, the genotype G3 and G4 would be considered more adapted to a wide range of environments than the rest of genotypes. Environments, such as Gazipur (E1) and Comilla (E2) could be regarded as a moderately stable site for high yielding hybrid rice improvement than other locations for grain yield due to IPCA score apparently near to zero which had little interaction effect. In AMMI 2 biplot, Comilla (E2), Barisal (E3) and Rangpur (E4) are the most discriminating environments, while Gan46A/ BRRI 10R (G2) and BRRI dhan28 (G5) are the most responsive genotypes. This indicates that analysis of multi-location trial data using AMMI model were more informative and important for determining visual comparisons, adaptability or stability in differentiating genotype response could be useful to plant breeders in supporting breeding program decisions.
Keywords: stability, adaptability, gxe and hybrid rice
Rice has a special significant position as a source of food providing over 75% of Asian’s staple food and more than three billion of world population’s meal which represents 50 to 80% of their daily calorie intake.1,2 This population will increase to over 4.6 billion by 2050.3 which will demand more than 50% of rice needs to be produced what is produced present to cope with the growing population.4,5 Yields of improved inbred rice varieties in favorable conditions have reached to a plateau or even subsequently declined in many countries including Bangladesh. It is recommended that a large number of high yielding rice with high adaption capability to diverse environments is required to accomplish specific socio-economic and agricultural needs. To meet the challenge of producing more rice under those constraints, we need new technologies like hybrid rice because it gives 15-30% yield advantage over inbred rice, increasing demand for hybrid rice especially in developing countries. Moreover, hybrid rice has also shown better performance under adverse conditions like drought and saline conditions. Therefore, multi-location trials need to be done before the hybrid rice promising genotype released as national varieties and passed on to farmers as end users. Varieties of hybrid rice developed in the plant breeding will eventually planted by farmers in various different environments. The results of multi-location trials of promising hybrid rice genotypes often reflects differences in grain yield in each location that the highest yield of a genotype in one location often showing is inconsistent in other locations. This is caused by the interaction between genotypes and environment, making it difficult for plant breeders in selecting the best genotype. Therefore, The G x E interaction is an important aspect of both plant breeding program and the introduction of new crop cultivars (Mc Laren and Chaudhary,6 Prasad and Sing7 and Freeman.8 Similarly, The AMMI model is a hybrid model involving both additive and multiplicative components of two way data structure which enabled a breeder to get precise prediction on genotypic potentiality and environmental influences on it. AMMI uses ordinary ANOVA to analyze the main effects (additive part) and principal component analysis (PCA) to analyze the non-additive residual left over by the ANOVA (Gauch17). The effectiveness of AMMI model has been clearly demonstrated by various authors using multilocation data in soybean,9 maize10 Wheat,11‒16 Field pea17. and rice,18 Das et al.,19 Tariku et al.,20; Nassir, 21; Islam et al.,22 Crop yield is a complex trait that is influenced by a number of component characters along with the environment directly or indirectly. Hence, if we could develop high yielding stable hybrid rice which adopted on diverse environments, to play a vital role in future food security of Bangladesh. The main objectives of the present study are to identify higher yielding promising hybrid rice genotypes having wide adaptation and/or specific adaptation to environment and environment interaction for their yield stability and adaptability across different environments.
Experimental design and plant materials
Multi environment trials on six hybrid rice genotypes were conducted during the 2009 boro cropping season at four environmental conditions; Gazipur (E1), Comilla (E2), Barisal (E3) and Rangpur (E4). The experimental materials consisting of 3 promising hybrid combinations (BRRI 9A/ BRRI 11R (G1),23 Gan46A/ BRRI 10R (G2) and BRRI 10A/ BRRI 11R (G3)) along with three checks (BRRI hybrid dhan2 (G4), BRRI dhan28 (G5) and BRRI dhan29 (G6) were planted in a plot of 4 m x 5 m. The experiment was laid out in a randomized complete block design with three replications. Thirty days old seedlings were transplanted in 20 square meter plot using single seedling per hill at a spacing of 20m2. Fertilizers were applied @ 270:130:120:70:10kg/ha Urea, TSP, MP, gypsum and ZnSO4, respectively. Standard agronomic practices were followed and plant protection measures were taken as required following the recommendation of Adhunik dhaner chash, BRRI.23 Two border rows were used to minimize the border effects. The grain yield (tha-1) data was collected at 14% moisture level.
Statistical analysis
The combined analysis of variance was proceeded to look at GxE and stability of the genotypes across all environments. The AMMI model, which combines standard analysis of variance with PC analysis,9 was used to investigate of G × E interaction. In AMMI model the contribution of each genotype and each environment to the GEI is assessed by use of the biplot graph display in which yield means are plotted against the scores of the IPCA.9
The AMMI model is:
Where, = yield of the genotype (g) in the environment (e); = grand mean; = genotype mean deviation; = environment mean deviation; N = No. of IPCAs (Interaction Principal Component Axis) retained in the model; = singular value for IPCA axis n; = genotype eigenvector values for IPCA axis n; = environment eigenvector values for IPCA axis n and = the residuals
Biplot analysis
Biplot analysis is the most powerful interpretive tool of AMMI models. Biplots are graphs where aspects of both genotypes and environments are plotted on the same axis so the inter-relationships can be visualised. There are two basic AMMI Biplots, the AMMI1 Biplot where the main effects (genotype mean and environments) are plotted against each other and the AMMI2 Biplot where scores for IPCA 1 and IPCA 2 are plotted.
Combined analysis of variance
The combined analysis of variance for grain yield of six rice genotypes is presented in Table 1. Genotype (G), environment (E) and genotype x environment interaction (GEI) were found highly significant (P<0.01) for grain yield. Effects of G and E showed highly significant mean sum of square (MSS) for grain yield was reflected genotypic differences towards adaptation to different environments, highly significant GxE effects suggests that genotypes may be selected for adaptation to specific environments, which is in harmony with the findings of Aina et al.,24 and Xu Fei-fei et al.,25 The significant G x E interaction effects demonstrated that genotypes responded differently to the variation in environmental conditions of location which indicated the necessity of testing rice varieties at multiple locations. This also shows the difficulties encountered by breeders in selecting new varieties for release. The factors explained SS (%) shows that rice grain yield was affected by genotype (54.86%) followed by environment (36.31%) and their interaction (6.27%).
Source |
df |
SS |
MSS |
Explained SS (%) |
Genotype (G) |
5 |
56.86 |
11.372*** |
54.86 |
Environment (E) |
3 |
37.63 |
12.545*** |
36.31 |
GxE interaction(GEI) |
15 |
6.5 |
0.433*** |
6.27 |
Error |
46 |
2.59 |
0.056 |
|
Total |
71 |
103.64 |
1.46 |
|
Table 1 Combined analysis of variance of grain yield for 6 rice genotypes evaluated at 4 environments
*** indicate significance at P<0.01 probability level; df, degree of freedom; SS, Sum of square; MSS, Mean sum of square
AMMI Analysis of variance
The AMMI analysis of variance for grain yield (tha-1) of six rice genotypes tested in four environments showed that 54.86% of the total sum of squares was attributed to genotypic effects, 36.31 % to environment effects and only 6.27% given by G x E interaction effects (Table 2). The large sum of squares values for genotypes indicated that the genotypes were diverse with large differences among genotypic means causing most of the variation in grain yield, which is accordance with the findings of Misra et al.,26 & Fentie et al.,27 in rice production. The presence of GEI was clearly demonstrated by the AMMI model, when the interaction was portioned among the first interaction principal component axis (IPCA) as they were significant P=0.01 in a postdictive assessment. The IPCA1 explained 5.81% of the interaction sum of squares with the degree of freedom 7. This implied that the interaction of the six rice genotypes at four environments was predicted by the first interaction principal components of genotypes and environments which is in line with the recommendation of Sivapalan et al.,28 for wheat. On the Other hand, the significant GxE interaction variance is suggestive of different performance of varieties under different environments.
Source |
df |
SS |
MS |
Explained SS (%) |
Genotype (G) |
5 |
18.954 |
3.791*** |
54.86 |
Environment (E) |
3 |
12.545 |
4.182*** |
36.31 |
GxE interaction(GEI) |
15 |
2.166 |
0.144*** |
6.27 |
IPCA1 |
7 |
2.007 |
0.287*** |
5.81 |
Error |
48 |
0.883 |
0.018 |
|
Total |
71 |
34.548 |
0.487 |
|
Table 2 Additive main effects and Multiplicative interaction (AMMI) analysis of variance for grain yield (tha-1) of 6 rice genotypes across 4 environments
*** indicate significances at the P<0.01 respectively.
Stability Analysis by AMMI model
The mean grain yield value of six rice genotypes over four environments presented in Table 3. It indicated that the genotypes G3 and G4 had the highest (7.43 tha-1 and7.84 tha-1) and the genotype G5 had the lowest (5.14 tha-1) productivity. Genotypes showed inconsistent performance across environments, G3 (7.43 tha-1) and G4 (7.84 tha-1) were the top performers, while G1 (6.78 tha-1) and G2 (6.74 tha-1) were moderate and G5 (5.14 tha-1) and G6 (6.00 tha-1) were the lower yielders. The environment mean grain yield ranged from 5.44 tha-1 to 7.13 tha-1 with an average of 6.65 tha-1. On the basis of environmental index value in terms of negative and positive value, E4 is poor and E1, E2 and E3 are rich environment. Within the genotypes G3 and G4 had higher and G1 and G2 were moderate average yield with positive index values which indicated that these genotypes might be adapted to favorable environments, while genotypes G5 and G6 might be adapted to poor environments. This result is in agreement with the findings of Islam et al., 22
Genotype/ Environment |
Gazipur (E1) |
Comilla (E2) |
Barisal (E3) |
Rangpur (E4) |
Genotype mean |
Index |
IPCA1 |
IPCA2 |
BRRI 9A/ BRRI 11R (G1) |
7.53 |
6.71 |
7.68 |
5.19 |
6.78 |
0.13 |
-0.47 |
0.19 |
Gan46A/ BRRI 10R(G2) |
7.36 |
6.69 |
8.01 |
4.92 |
6.74 |
0.09 |
-0.77 |
0.02 |
BRRI 10A/BRRI 11R(G3) |
7.93 |
7.72 |
7.76 |
6.29 |
7.43 |
0.77 |
0.22 |
-0.27 |
BRRI hybrid dhan2 (G4) |
8.2 |
8.08 |
8.52 |
6.55 |
7.84 |
1.18 |
-0.04 |
-0.26 |
BRRI dhan28 (G5) |
5.48 |
5.22 |
5.43 |
4.42 |
5.14 |
-1.52 |
0.48 |
0.33 |
BRRI dhan29 (G6) |
6.28 |
6.27 |
6.19 |
5.27 |
6 |
-0.65 |
0.57 |
-0.01 |
Environment mean |
7.13 |
6.78 |
7.26 |
5.44 |
GM=6.65 |
|||
Index |
0.88 |
0.13 |
0.61 |
-1.21 |
||||
IPCA1 |
-0.28 |
0.23 |
-0.77 |
0.83 |
||||
IPCA2 |
0.13 |
-0.46 |
0.08 |
0.24 |
||||
SE |
0.07 |
0.05 |
0.13 |
0.22 |
||||
CV (%) |
2 |
1 |
3 |
7 |
||||
5% LSD |
0.22 |
0.15 |
0.4 |
0.69 |
|
|
|
|
Table 3 Stability analysis for grain yield (t/ha) of 6 rice genotypes in 4 environments
AMMI 1 biplot display
The AMMI 1 biplot gave a model fit 99.5% (Figure 1). This result is in agreement with the findings of Naveed et al.,29; Gauch et al.,30 and Annichiarico.31 Genotypes and environments on the same parallel line, relative or ordinate have similar yields and a genotype or environment on the right side of the midpoint of this axis has higher yield than those of left hand side. Consequently, among the genotypes, G3 and G4 were generally exhibited high yield with small IPCA scores but the genotype, G3 being the overall best with showing positive IPCA1 score near zero and the genotype G4 showed negative IPCA1 score very close to zero indicating that these varieties were stable and less influenced by the environments Yau SK32 Similar outcomes have reported by Das et al.,33 and Kulsum et al.,34 Hence, the genotype G3 was identified as specially adapted culture to the environments E2. The genotypes G1, G2, and G4 were considered as the favorable environments for E1 but the genotypes G2 had large negative IPCA score which indicated higher interaction with environments thus they are unstable and genotypes G5 and G6 were showed positive IPCA1 score with below average yield and these two genotypes were favorable environments for E4. On the other hand, regardless of the positive or negative values, the environment E3 and E4 had large IPCA scores which indicated these environments are unstable and higher interaction with genotypes but the other two environments E1 and E2 showed small positive IPCA score apparently near zero with high mean value and hence had small interaction effects indicating that all the genotypes performed well in these locations. This result is an agreement with the findings of Adugna et al.,36 Anandan et al.,38 and Islam et al.,22
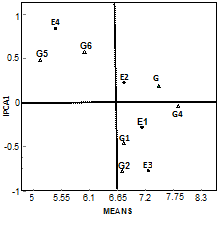
Figure 1 AMMI 1 Biplot for grain yield (tha-1) of six rice genotypes (G) and four environments (E) using genotypic and environmental scores
AMMI 2 biplot display
In AMMI 2 biplot, the environmental scores are joined to the origin by side lines. Sites with short spokes do not exert strong interactive forces. Those with long spokes exert strong interaction. An example of this is shown in Figure 2 where the points representing the environments E1, E2, E3, and E4 are connected to the origin. The environments E1 had short spokes and they do not exert strong interactive forces but the environments E2, E3 and E4 had long spokes they are exert the most discriminating environments in Figure 2. Hence, the genotypes near the origin are not sensitive to environmental interaction and those distant from the origins are sensitive and have large interaction. In this case, the genotypes, G2 and G5 had more responsive since they were away from the origin. The multivariate approach, the AMMI model is better for partitioning the GxE into the causes of variation, which easier identify environments’ potential and used to identify superior genotypes either specific adaptation. Similar result was reported by Anandan et al.,36 ; Crossa37 and Kempton.38
From the present investigation it is concluded that stability analysis provided a good understanding of the adaptation level of rice genotypes across a diverse range of environments. The yield stability across different environment varied among genotypes. AMMI statistical model could be a great tool to select the most suitable and stable high yielding hybrids for specific as well as for diverse environments. As a result, almost all of the evaluated genotypes were affected by the genotype x environment interaction effects, so that no genotype had superior performance in all environments. In this study, the combined variance analysis of variance indicated that the genotypes (G), environments (E) and GxE interaction were significant at P<0.01 level. The mean grain yield value of genotypes over environments indicated that G3 and G4 had the highest (7.43tha-1 and 7.84 tha-1) and G5 had the lowest yield (5.14 tha-1), respectively. It is noted that the variety G4 showed higher grain yield than all other varieties over all the environments. Most of the genotypes showed environment specificity. The AMMI 1- biplot model classified the testing environments into three sections. According, one of the tested genotypes G3 were found to be best for environment E2 and three genotypes G1,G2 and G4 were found to best fit for environment E1 and E3, while the other two genotypes G5 and G6 were found to best fit for environment E4. The AMMI-2 biplot showed that the genotypes G1 and G2 with respect to sites E1 and E3 and the other two environments, E2 and E4 were well fit for genotypes G3, G6 and G5 while the genotype G4 was not found to be fit to any of the testing locations.
Authors are thankful to all the staffs of Hybrid Rice Research & Development GOB project funded by Ministry of Agriculture and the yield trials conducted in different locations was supported by late Dr. A.W Julfiquar of Director Admin and Project Director of the Project, Bangladesh Rice Research Institute, Bangladesh, Gazipur -1701 and also express thanks to M R Islam for helping AMMI biplot statistical analysis.

©2019 Akter, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.