Advances in
eISSN: 2373-6402


Review Article Volume 2 Issue 1
1Institute of Soil, Water and Environmental Sciences, Gilat Research Center, Israel
2Agronomy Department, Institute of Food and Agricultural Sciences, University of Florida, USA
Correspondence: Ratna Karan, Department of Agronomy, Institute of Food and Agricultural Sciences, University of Florida, Gainesville-32611, Florida, USA, Tel +1-225-612-1964
Received: November 01, 2014 | Published: January 24, 2015
Citation: Soda N, Wallace S, Karan R. Omics study for abiotic stress responses in plants. Adv Plants Agric Res. 2015;2(1):28-34. DOI: 10.15406/apar.2015.02.00037
Abiotic stresses, such as drought, temperature extremes and salinity, are the major constraints on crop yield and quality in fields. TranscriptOmics, proteOmics and metabolOmics have been employed to improve understanding of the biological processes and molecular/cellular mechanisms involved in plant stress responses. Over the last several decades, in the light of research carried out using different Omics approaches, various stress related mechanisms have been proposed for the development of tolerant varieties. Integrated use of functional genOmics helps in understanding the relationship between an organism’s genome and their phenotype under different environmental conditions. By exploiting available genetic information and continuously improving techniques and strategies, integrated functional genOmics alongside bioinformatics will provide a foundation for further in–depth functional studies of stress tolerance in plants.
Keywords: abiotic stress, metabolomics, plants, proteomics, transcriptomics
NGS, next generation sequencing; NIPGR, national institute of plant genome research; CTDB, chickpea transcriptome database; SSH, substractive suppression hybridization; mRNA, messenger RNA; PTMs, post–translational modifications; GABA, gamma amino butyric acid; TCA, tricarboxylic acid; BCAAs, branched chain amino acids; SAGE, serial analysis of gene expression
In fields, plants are frequently subjected to various environmental stresses such as water deficit, freezing, heat and salt stress. These environmental stresses responsible for the large gap existing between the potential yield and real and harvest yield in several crops worldwide.1 As plants are sessile in nature, they have developed different strategies to adapt and grow under rapidly changing environments. These strategies involve rearrangements at the molecular level, during gene expression starting from transcriptional regulation to mRNA processing, translation, protein modification or its turnover. Under stress condition, plants show stress specific regulation of transcription that affects their transcriptome.2–5 These transcriptionally regulated genes have different functions, such as transcription, translation, signaling, metabolism and general stress response. Generally, seedling and reproductive stages are more susceptible to stress. Thus, stress response studies during these growth stages reveal novel differentially regulated genes or proteins with important functions in plant stress adaptation. In the past few decades, a great deal of research has been conducted with regards to deciphering the mechanism of multi–stress tolerance in crop plants. A large number of candidate genes, proteins and pathways have been identified using Omics approaches (Figure 1). Omics refers to a study of large sets of molecules in biology, for detection of genes (genOmics), mRNA (transcriptOmics), protein (proteOmics), and metabolites (metabolOmics). Though plant biologists have unveiled several stress related molecular mechanisms, we are still a step away from understanding the plants entire response to multiple stresses in fields. While advancement in microarray and deep–sequencing technologies lead to rapid accumulation of genomic and transcriptomic data under different abiotic stresses,6,7 the limited depth of quantitative proteOmics has inhibited similar progress in post–transcriptional gene regulation. To date, transcript levels are routinely used as the only measure for gene expression in high–through put approaches. Several studies, however, have reported a low correlation between transcript and protein levels, highlighting the importance of post–transcriptional processes as the limited predictive value of transcripts for protein expression.8–12 Recent developments in research technologies have shown that understanding of plant stress responses using genOmics, transcriptOmics, proteOmics and metabolOmics studies, requires quantitative information at every step of gene expression. Several research groups have shown the relevance of post–transcriptional and translational regulatory mechanisms in the plant adaptation process to different abiotic threats, including light,13,14 heavy metal intoxication,15 dehydration16,17 and salinity stress.18
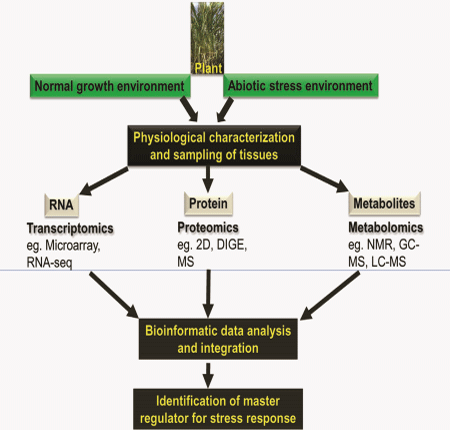
Despite all the recent developments, our understanding of post–transcriptional gene regulation and its effects on protein–complex stoichiometry are lagging behind. Transcriptomic technologies have advanced tremendously in recent years. RNA–seq has shown to be an excellent method to obtain unbiased transcriptome data that correlate well with micro array analyses.19 Though not as advanced as transcriptomic techniques, the mass spectrometry methods for high–through put analyses of proteome are also advancing rapidly. However, whilst these technologies are currently applied to answer many research questions in various model systems, there are relatively few comparative studies involving these technologies. Complementary analysis of the proteome and metabolome combined with a comprehensive transcriptome provides an important validation tool for the expression of key genes. Dissecting the relationship between differentially expressed transcriptome, proteome and metabolome will potentially help us to understand the molecular mechanisms underpinning abiotic stress tolerance in plants. By investigating transcript–protein–metabolite correlations, and change of correlation between the normal and stressed state, we can identify biological processes that are strongly regulated by abiotic stresses in plants. Furthermore, these studies will provide a comprehensive data set for quantitative systems–level analyses.
Transcriptomics: A key to understanding abiotic stress responses in plants
Transcriptomic approaches have paved the way to understanding plant responses to abiotic stresses. Recently, transcriptome analysis by next generation sequencing (NGS), RNA–seq for sRNAs and their use in genOmics research, have greatly improved plant genomic resources.20–25 Li et al.,26 identified 5365 differentially expressed probe sets (2–fold as cutoff) in the switch grass cultivar Alamo, under heat stress, utilizing switch grass Affy metrix gene chips. By comparative transcriptome analysis in response to heat stress, they identified 16 common genes in four monocots–switch grass, rice, wheat and maize. Interestingly, most of them were associated with protein refolding processes; therefore, these genes can be used as valuable biomarkers for identifying heat sensitive plant germplasm. Wakasa et al.,27 have done RNA sequencing–mediated expression profiling of transgenic rice plants, produced by homologous recombination, in which endogenous genomic OsIRE1 (ER stress sensor/transducer) was replaced by missense alleles defective in ribonuclease activity. This study provided valuable information about the ER stress response in rice plants and led to the discovery of new genes related to ER stress. Instead of just one type of stress study that is generally conducted under laboratory conditions, plants grown in the field are exposed to a combination of stresses, which require agonistic or antagonistic responses, or a number of potentially unrelated responses to a certain single stress condition. Rasmussen et al.,28 analyzed such responses by comparing transcriptome changes in 10 Arabidopsis thaliana ecotypes under different stresses and their combinations. This study revealed that 61% of the transcriptome changes in response to double stresses were not predicted from the responses to single stress treatments. They have also shown that plants prioritized between potentially antagonistic responses for only 5% to 10% of the responding transcripts. In light of this research, authors have delineated co expression network modules responding to single and combined stresses. RNA–seq analysis of Chenopodium quinoa under four water treatments (field capacity to drought) showed an overlap between drought stress tolerance and other abiotic stress mechanisms.29
Kudapa et al.30 used several Sanger EST collections of chickpea, together with sequence data from two different NGS (Next Generation Sequencing) platforms (Illumina and FLX/454) of chickpea, to produce a more extensive chickpea transcriptome assembly (CaTA v2). Additionally, NIPGR (National Institute of Plant Genome Research, India) have developed the Chickpea Transcriptome Database (CTDB), which will provide comprehensive information about the chickpea transcriptome (http://www.nipgr.res.in/ctdb.html). Apart from NGS, substractive cDNA suppression hybridization (SSH) technology has also proved to be very helpful in transcriptomic studies in unveiling stress responses.31 Molina et al.,6,7 used high resolution power of Super SAGE (Serial Analysis of Gene Expression) coupled to the Roche 454 Life/APG GS FLX Titanium NGS technology, to characterize the complete transcriptome of drought and salt–stressed chickpea plant’s roots and nodules under stress condition.
Transcriptome sequencing of chrysanthemum plants under dehydration stress using the Illumina sequencing also provided better understanding of the molecular mechanisms of dehydration stress responses.32 Zhu et al.,33 applied a comparative microarray analysis approach to study the transcriptome changes of cotton under five abiotic stresses. Their study unveiled the functional genes and stress related pathways, and also suggested a crosstalk of responsive genes or pathways to multiple abiotic stresses in cotton seedlings.
Transcriptomic technologies have the potential to provide deep coverage and unbiased representation of transcript abundance, which is very important in non–model plants lacking genome sequence information.34 However, the frequent incongruity between protein levels and the abundance of cognate gene transcripts suggested the need of complementary analysis of the proteome for further validation of candidate genes and pathways.35
Proteomics: a closer look at translatome regulating cellular responses
Most functional genOmics studies rely on transcriptome analysis across changing external conditions to identify transcripts that are differentially regulated, and often it is hypothesized that changes in transcript levels may lead to corresponding changes in protein levels. However, it has been shown by several groups that protein levels do not always necessarily corresponds to mRNA levels.36,37 Anderson et al.,38 reported that the correlation coefficient in quantity between mRNA and protein abundance is relatively low. Protein level can be modulated by changing either the rate of synthesis or the stability of the messenger RNA (mRNA or transcript), or the synthesis or stability of the protein itself.39 Translatome refers to the pool of all RNAs that are associated with ribosomes purified via an affinity tag. Study of the translatome or proteome can confirm the presence of the protein, and provides a direct measure of the quantity present. Precise analysis of the translatome/proteome and metabolome is essential for understanding the fundamentals of stress physiology and biochemistry. Proteins, the functional translated portion of the genome, play an essential role in plant stress response. Proteomic studies provide us with a finer picture of the protein networks and metabolic pathways primarily involved in stress tolerance mechanism. Identifying master regulator proteins that play key roles in the abiotic stress response pathway is fundamental in providing opportunities for developing genetically engineered crop plants to allow us to understand the stress response. Halbeisen et al.,40 compared the transcriptome with the translatome in yeast cells, exposed to different stresses, to determine the discrepancy between transcript and protein levels. Their analysis suggested that transcriptome and translatome show a strong coordinated response, particularly under severe stress. While, under mild stress conditions about 2% of all expressed messages showed differential regulation, and therefore represent candidates for translational regulation. In A. thaliana, precise mapping of ribosome footprints (RFs) on mRNAs to investigate translational regulation under control and sublethal hypoxia stress conditions, demonstrated nearly 100–fold variation in the efficiency of translation of individual mRNAs under both conditions. These findings therefore provide unique insights into posttranscriptional and translational regulation modulated by lower level of applied stress.41 Yanguez et al.,42 carried out a genome–wide analysis to monitor the changes in the translation efficiency of individual mRNAs of A. thaliana seedlings after exposure to temperature stress. They demonstrated that, translation exerts a wide regulation on gene expression. While for some mRNAs translation is severely repressed, translation of homeostasis and stress related mRNAs follow a differential pattern. Their study suggested that, mRNAs with special features, such low 5′–UTR G+C content and small cDNA length, are preferentially translated.
Several research groups have extensively explored proteOmics tools to solve the maze of heavy metal ion stress responses in plants. Under Cd stress, in Brassica juncea L. roots, over expression of sulfite reductase and O–acetyl serine sulfhydrylase proteins revealed the reduction of sulfate to cysteine.43 A leaf mesophyll tonoplast proteOmics study involving Hordeumvulgare L, reveals an MRP–like ABC transporter and two novel CAX transporters (CAX1a and CAX5), assuring Cd2+ transport into the vacuole.44 Additionally, in Glycine max L. leaves, abundance of Hsp70 and peroxiredoxin were reported,45 and up–regulation of proteins associated with Cd–chelating pathways were reported in different plant species viz. Linumusitatissimum, A. thaliana and G. max.46–48 Similar research has been carried out to decipher cellular responses towards other heavy metals, such as with B deficiency in Lupinus albus roots, where proteins involved in cell division and metabolic processes were found to be down regulated.49 Under Cr stress, enhanced expression of proteins involved in ROS detoxification, defense responses, photosynthesis and chloroplast organization were reported in Zea mays.50 Accumulation of Cr–responsive proteins linked to heavy metal tolerance and senescence pathways were reported in Miscanthus sinensis.51 Furthermore, in the presence of Al, the stress tolerant genotype of G. max showed accumulation of enzymes which catalyze synthesis of citrate (involved in Al3+ detoxification) whereas, the sensitive genotype showed induction of proteins related to general stress response.52
Subba et al.,53 studied the nuclear proteome in two contrasting chickpea cultivars under drought stress, to identify the proteins playing key roles in drought stress tolerance. Researchers are also exploiting a2DE (two dimensional gel electrophoresis) approach for generating a comprehensive nuclear54,55 and cell wall proteome,56,57 which will provide a basis for future comparative studies. In addition, Heidarv et al.,58 studied time course dynamics of physio–biochemical and proteome changes in chickpea under cold stress. With this comparative study of biochemical and molecular events under cold stress, they have provided a more comprehensive view of chickpea stress responses.
Besides differential protein abundance, post–translational modifications (PTMs) also increase protein complexity and dynamics, regulating different cellular events. Advances in proteOmics techniques made global identification of PTMs feasible. Patton59 described thesimple and specific methods, suitable for different PTMs, for visualization of modified proteins in a gel. PTM studies are further aided by the sensitivity of mass spectrometry, to help in analyzing the in vivo phosphorylation of proteins at the proteome scale.60 For example, in salt–treated rice roots, increased relative abundance of 17 phosphoproteins (e.g., dnaK–type chaperone HSP70, putative GST, small GTP–binding protein OsRac2, mannose–binding RICE lectin), and a decreased relative abundance of 11 phosphoproteins (e.g., putative protein kinase, ATP synthase β chain) have been reported. Tanou et al.,61 suggested an important role of protein carbonylation and S–nitrosylation patterns in the salinity response of citrus. Their study has shown a strong increase in the level of carbonylated proteins (ADH, chaperonin 60 subunit α, glycolytic enzymes, HSP70, RubisCO LSU, subunits of chloroplast and mitochondrial ATP synthase F1, mitochondrial processing peptidase) and nitrosylated proteins (actin, annexin, ENO, GAPDH, GST, HSPs, RubisCo activase, RubisCo LSU, PGK, TPI, SOD, peroxiredoxin, tubulin, several eIF and eEF) under salt stress. Kosova et al.,62 also reported salinity related PTMs in proteins and the effect they have on protein function and catalytic activity. Despite the recent rapid developments in the field of transcriptOmics and proteOmics, a great deal of investigation of biochemical regulation was required before a thorough understanding of plant stress responses could be reached. This gap in understanding led to the development of metabolOmics approaches.
Metabolomics: pinning down the stress responsive pathways
GenOmics, transcriptOmics, and proteOmics help in identifying the candidate genes and proteins playing crucial roles in plant stress responses. However, the delineation of the regulatory networks and metabolic pathways responding to single and multiple concurrent stresses is required for better understanding stress response in crop plants. In the past few years, metabolOmics provided a new zenith to plant stress related studies, and became an indispensable tool in understanding the molecular mechanisms underlying stress responses.63 On exposure to any stresses, the plant metabolism undergoes reconfiguration in order to maintain the metabolic homeostasis and production of compounds that ameliorate the stress. In light of recent advancements, such as metabolOmics and systems biology approaches, detailed information regarding crucial components of plant metabolic pathways has been obtained. Metabolomic studies of A. thaliana plants subjected to drought stress revealed the accumulation of several metabolites, including amino acids such as proline, raffinose family oligosaccharides, gamma amino butyrate (GABA), and tricarboxylic acid (TCA) cycle metabolites, which are known to respond to drought stress in plants.64 With the help of mutant and transcriptomic approach, ABA dependent transcriptional regulation responsible for activation of most of the stress related metabolic pathways have been demonstrated.64 Metabolite profiling also revealed temporal distinction in A. thaliana leaves under mild osmotic stress.65 Mature leaves showed typical drought responses, such as accumulation of proline, erythritol and putrecine, while many of these metabolites decreased in expanding leaves, which correlates with the transcriptional response.65 Metabolic studies also highlighted the variable response of plant metabolism in different developmental stages and degrees of desiccation. In the case of amino acid metabolism, most amino acids were accumulated in severely desiccated leaves but decreased in mildly desiccated leaves. Metabolite profiling of maize and wheat, exposed to water stress conditions, suggested common changes in the levels of metabolites including branched chain amino acids (BCAAs).66,67 Verslues and Juenger68 revealed an important role of metabolic regulation, including regulation of photosynthesis and accumulation of osmolytes during drought stress response in A. thaliana. Caldana et al.,69 carried out transcriptome and metabolome profiling of A. thaliana under eight environmental conditions. Metabolic response to high light showed accumulation of the photo respiratory intermediates, glycine and glycolate in the early phase. Interestingly, they have reported similar responses during the mid–phase of high light stress and low temperature treatments, including accumulation of shikimate, phenylalanine and fructose, and the decrease of succinate; however, the physiological meaning of these overlapped responses is currently not known.
Kusano et al.,70 studied the effect of UV light on A. thaliana metabolism and documented a biphasic response. In the early phase, they reported major changes in the levels of primary metabolites, including ascorbate derivatives. By contrast, mid–to late–term responses were observed in the classically defined UV–B protectants, such as flavonoids and phenolics. Their results suggested cell priming upon early exposure to UV–B, which involves reprogramming of the metabolism for efficient diversion of carbon towards aromatic amino acid precursors of the phenyl propanoid pathway. They have also suggested the importance of ascorbate in the short–term response to UV–B. Furthermore, this group combined transcriptOmics with metabolOmics to determine the metabolic changes responsible for adaptation to increased exposure to UV–B, and metabolic changes involved in the perception–signaling relay, which alerts the plant cell to respond against the stress.71 Metabolite profiling of A. thaliana leaves also helped in elucidating the metabolic basis of dark–induced senescence and function of the mitochondrial alternative electron transport pathway during dark treatment.72 In other metabolite profiling studies, an increase in BCAAs, i.e.valine, leucine, isoleucine, and other amino acids sharing synthetic pathways with BCAAs i.e. lysine, threonine and methionine were reported under abiotic stress conditions.72,73 The authors have suggested that BCAAs function as compatible osmolytes, since drought stress led to increased accumulation of BCAAs in various plant tissues. Interestingly, protein degradation serves as an alternative respiratory substrate for stressed plants.74
Although, all of these approaches greatly increased our knowledge with regards to candidate genes, proteins and pathways playing crucial roles in plant stress responses, there is still a long avenue to explore. Recently, researchers have combined either two or all three Omics approaches to obtain a holistic view of stress responses.35,75–79 Mehmeti et al.,80 reported discrepancies between transcriptomic, proteomic and metabolite data of lactic acid bacteria, suggesting regulation beyond the level of transcription. These discrepancies are either because of noise/poor statistics for the microarray data or due to varying efficiency of transcription and translation. They have indicated additional posttranscriptional or translational regulation in these bacterial cells. Complementary analysis of the proteome and metabolome combined with a comprehensive transcriptome provided an important validation tool for the expression of key genesin Macleaya sp.79 Combinations of transcriptome, proteome and metabolite profiling helps in revealing the correlation between the expression of stress related biosynthetic genes and corresponding metabolic products, which further increases our understanding of plant stress responses.75,78 Zeng et al.,79 also combined these Omics approaches to reveal alkaloid biosynthesis in Macleaya sp. While studying phosphate–deficient A. thaliana roots, Lan et al.,81 revealed multiple levels of gene regulation, and suggested integrated measurement and interpretation of changes in protein and transcript abundance for generating a complete inventory of the components that are critical for stress responses. Colmsee et al.,82 established a data warehouse for maize, OPTIMAS–DW. It can handle different data domains, integrates data from different data domains, such as transcriptOmics, metabolOmics, ionOmics, proteOmics, phenOmics, and enables the user to find answers to different systems biology questions. Amiour et al.,83 discussed the potential use of ‘Omics’ studies to improve understanding of whole plant nitrogen econOmics in maize.
Srivastava et al.,84 proposed a data evaluation strategy to provide an efficient way of compiling complex, multi–platform datasets to obtain significant biological information. This study of transgenic populus plants harboring superoxide dismutase gene, provided system–level information on ROS metabolism and responses to oxidative stress. Yang et al.,85 also reviewed applications of Omics approaches for understanding secondary metabolism, including the discovery of novel genes, the identification of gene function, and the detection of novel pathways of the metabolic network. All of these studies accelerate our understanding of plants interaction with their environment, and their performance under prevailing stress conditions.
Plants are the primary producers on earth and abiotic stresses affect their growth, development and final yield potential. Under stress conditions, plants modulate themselves to transiently adapt to the existing circumstances by changing the expression pattern of genes, proteins and metabolites. To identify those changes, various tools and techniques, such as genOmics, transcriptOmics, metabolOmics, ionOmics and phenOmics, have been devised to allow us to better understand the genetic makeup of plants and their adaptability potential under stress conditions. Recent Omics studies have accumulated a great deal of information at transcript, protein and metabolite levels to perceive the survival potential of plants under stress. However, a highly coordinated approach, such as system biology, is needed for the full comprehension of the complex regulatory nature of plants so that master stress regulators can be identified.
None.
The author declares no conflict of interest.

©2015 Soda, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.