Advances in
eISSN: 2373-6402


Research Article Volume 3 Issue 4
Kiomars Rouhrazi, Department of Agriculture, Malekan branch, Islamic Azad University, Malekan, Iran
Correspondence: Department of Agriculture, Malekan branch, Islamic Azad University, Malekan, Iran, Fax 984-137-8426-79
Received: April 11, 2016 | Published: April 27, 2016
Citation: Azizpour N, Rouhrazi K. Isolation and characterization of rhizosphere bacteria for the biocontrol of the asochyta rabiei in Iran. Adv Plants Agric Res. 2016;3(4):121-125. DOI: 10.15406/apar.2016.03.00104
Chickpea (Cicer arietinum), a cool season grain legume grown on a surface of 11million ha with worldwide production of about 9 million tons, is cultivated in more than 45 countries throughout the world including Iran where it is grown mostly in rainfed areas and on marginal lands in the province of West Azarbayjan. Ascochyta blight is perhaps the most frequent and damaging disease of chickpea worldwide. It is caused by Ascochyta Rabiei; a fungus that selectively attacks chickpea. Ten isolated bacteria from chickpea plants rhizosphere soil, were evaluated in vitro as a potential antagonist of fungal pathogen. Sequencing of 16S rDNA and comparison with Gen Bank database of sequences revealed that antagonistic strains belong to the species Pseudomonas fluorescens (three strains), P. putida (three strains), Bulkholderia multivorans (three strains) and Mezorhizobium ciceri (one strain). All the strains significantly inhibited A. rabiei, and resulted in >30% inhibition on PDA.
Keywords: antagonistic bacteria, aschochyta rabiei, chickpea, fungal pathogen, antagonistic strains, cicer arietinu, ascospores, grain legume, rhizospher bacteria, cellulose, chitinase
AR, Ascochyta Rabiei; PGPR, plant growth-promoting rhizobacteria; BCA, biocontrol agents; NA, nutrient agar
Chickpea (Cicer arietinum), a cool season grain legume grown on a surface of 11 million ha with worldwide production of about 9 million tons,1 is cultivated in more than 45 countries throughout the world including Iran where it is grown mostly in rainfed areas and on marginal lands in the province of West Azarbayjan. Ascochyta blight is perhaps the most frequent and damaging disease of chickpea worldwide. It is caused by Ascochyta rabiei (AR), a fungus that selectively attacks chickpea. AR survives in plant debris, soil and infected seeds and it reproduces asexually, through the production of conidia in flask-shaped pycnidia, and sexually, through the production of ascospores from pseudothecia. Ascospores and conidia spread through wind and rain splashes, travelling distances up to hundreds of meters.2
Infections may arise from seed borne inoculum or from windborne spores (ascospores). Infections usually begin low in the crop canopy during periods of cool, wet weather. All parts of the plant above the soil line are subject to attack and may develop elongated, sunken, dark lesions. Under cool moist conditions, the patches of diseased plants in the field may rapidly increase in size, and lesions may develop higher in the crop canopy on leaves and pods. Such seed contamination is not always visible nor is the fungus in or on the surface of seed easy to detect in the laboratory. Only heavily infected seeds will bear visible blight symptoms, which include small size wrinkles, lesions, and/or dark discoloration.3
The control of the soil-borne pathogens is difficult because of their ecological behavior, their extremely broad host range and the high survival rate of resistant forms, under different environmental conditions. Many research studies have shown that biological control offers an environmentally friendly alternative to protect plants from soil-borne pathogens.4-6 Although the number of biocontrol products is increasing, these products still represent only a very small proportion of fungicides.7 In recent years, several bacterial and fungal antagonists against soil-borne plant pathogenic fungi have been described.8,9 Application of biocontrol agents (BCAs) or plant growth-promoting rhizobacteria (PGPR) is considered as an important approach in crop protection against plant pathogens. Several microbes have been studied extensively as BCAs against various phytopathogens and these also showed plant growth promotion activity.10,11 The objectives of this study were to select and characterize rhizospher bacteria with antagonistic activities against A. rabiei.
Fungal material
The isolates of AR used in this study were obtained by isolation from samples of stems, sheets and chickpea pods presenting of the symptoms of Ascochyta blight.
Isolation of antagonistic bacteria
For isolation of bacteria, roots of healthy chickpea were excised and placed into 10 mL of a sterile 0.9% NaCl solution and vortexed for 10 min in order to detach the associated rhizosphere soil. Serial dilutions of the resulting root wash were plated on nutrient agar (NA) supplemented with ampicillin (100 mg/mL).12 Plates were incubated at 28 8C for 24-48 h. To obtain the most abundant bacteria from each sample, selection of strains with different colony morphology was performed from the highest dilutions. The isolates were stored in 30% (v/v) glycerol solution at -80°C.
Evaluation of strains for in vitro biological control
Isolated bacteria from rhizosphere were tested against AR in plate bioassays. AR were cultivated in PDA at 28 8C. Conidia were harvested from the surface of plates by flooding the 10-day-old cultures with 9 mL of sterilized distilled water and gently scraping with a sterilized glass rod; conidial concentration was determined with a Neubauer chamber.13 Plates containing the media to be tested (NA, PDA) were prepared. Then, an agar over-layer containing the target fungus, immobilized at a concentration of 104-105 conidia/mL, was placed on the medium. The methodology described by Montesinos et al.14 was followed in order to prepare the overlay, using 0.7% agar. Four milliliters of the medium were placed in screw-capped test tubes, which, once sterilized, were kept inside a water bath at 35 0C. Next, 10 mL of a target conidia suspension were added to each test tube, which were vortexed; the content of each tube was then homogeneously distributed on a plate containing the same culture medium. The bacterial strains tested were sown by gently touching the agar surface with a sterile toothpick, previously inoculated by touching the surface of a single colony. Plates were incubated for 72 h at 28 8C. The degree of inhibition in each medium was determined by measuring the halo around the bacterial strain without fungal growth. Three replicates were considered for the value of the inhibition halo.
Identification of antagonistic bacteria
Different phenotypic features were assessed, including the Gram stain, oxidase test, production of acids from 1% glucose in oxidation/fermentation (OF) basal medium and utilization of sole carbon and nitrogen sources. Molecular identification of antagonistic strains that were used in this study was made by sequencing the 16S rRNA gene. Amplification was carried out by PCR with fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and rD1 (5′-AAGGAGGTGATCCAGCCGCA-3) designed by Weisburg et al.15 Standard PCR conditions were 1 min DNA denaturation at 94 0C, 1 min annealing at 57 0C, and 1 min extension at 72 0C for 35 cycles. Multiple sequence alignment of 16S rDNA sequences of antagonistic strains and those of related bacteria was performed with the Clustal W program of DDBJ. An evolutionary tree for the datasets was inferred by the neighbor-joining (NJ) method of Saitou and Nei16 using the NJ program of Molecular Evolutionary Genetics Analysis version 5 (MEGA5). The stability of relationships was assessed by performing bootstrap analysis of the NJ data based on 1000 re samplings.
Production of hydrolytic enzymes
Proteolytic activity was detected by inoculating the strains on a medium composed of 1% casein and 2.3% agar dissolved in Castan˜eda medium.17 Plates were incubated for 48 h at 28 8C. Casein hydrolysis was detected by the formation of a whitish (opaque halo coagulated casein) around a translucent area (totally hydrolyzed casein), surrounding the colony. To determine the cellulolytic activity, carboxyl methyl cellulose (CMC) was incorporated at 0.1% into the YEMA-0.2% mannitol agar plates. Colonies were grown for three days at 28 0C and washed off with water. The plates were then flooded with 0.1% (wt:vol) Congo Red in water for 15 min, washed for 10 min with 1 M NaCl, and then washed for 5 min with 5% acetic acid. Degradation of CMC was observed as clearings (reduction of staining). All the hydrolytic tests were performed twice.
Assay of chitinases
strains were cultured at 28 8C for 96 h on a rotary shaker in 250 mL conical flasks containing 50 mL of chitin–peptone medium (glucose 0.5%, peptone 0.2%, colloidal chitin 0.2%, K2HPO4 0.1%, MgSO4_7H2O 0.05%, and NaCl 0.05%, pH 6.8). The cultures were centrifuged at 12,000 g for 20 min at 4 0C and the supernatant was used as enzyme source. Colloidal chitin was prepared from crab shell chitin according to Berger and Reynolds.18 The reaction mixture contained 0.25 mL of enzyme solution, 0.3 mL of 1 M sodium acetate buffer (pH 5.3) and 0.5 mL of colloidal chitin (0.1%). The reaction mixture was incubated at 50 0C for 4 h in a water bath. Chitinase activity was determined by measuring the release of reducing sugars by the method of Nelson.19 One unit of chitinase was determined as 1 nmol of GlcNAc released per minute per mg of protein. Protein content in all the samples was determined as described by Bradford using bovine serum albumin as the standard.
Ten bacterial strains were obtained from chickpea roots. Recovered bacterial strains were tested for their antagonistic ability against the phytopathogenic fungi AR. Result of antagonistic activity of the strains against AR showed in Table 1. Sequencing of 16S rDNA and comparison with Gen Bank database of sequences revealed that antagonistic strains belong to the species Pseudomonas fluorescens (three strains), P. putida (three strains), Bulkholderia multivorans ( three strains) and Mezorhizobium ciceri(one strain). All the tested strains resulted in > 30% inhibition on PDA. The result of Effect of strains on the mycelial growth of AR showed in Table 1. Five strains (A1, A3, A4, A5 and A6) showed protease activity. Three strains (A2, A8 and A10) produced cellulase or chitinase. Additionally, seven strains (A1, A2, A3, A4, A5, A6, A7 and A8) were able producing siderophores in CAS medium.
Bacterial strain |
Inhibition percentage of mycelium growth |
A1 (Pseudomonas fluorescens) |
18.23a |
A2 (P. fluorescens) |
16.5b |
A3 (P. fluorescens) |
16.24b |
A4 (P. putida) |
12.74c |
A5 (P. putida) |
17.62b |
A6 (P. putida) |
13.20c |
A7 (Bulkhorderia multivorans) |
11.25c |
A8 (B. multivorans) |
11.10c |
A9 (B. multivorans) |
12.11c |
A10 (Mezorhizobium ciceri) |
5.2d |
Table 1 Effect of bacterial isolates on Ascochyta Rabiei , measured as diameters of the fungal colonies in the presence of each antagonistic bacterial strain, compared to the control plate
Among the fungal diseases that affect chickpeas around the world, one of the most serious is the Ascochyta blight, caused by AR. Ascochyta blight is a highly destructive disease in most chickpea-growing areas of the world.20,21 The pathogen grows asexually on the host plant, while its perfect stage can be recovered from wintering chickpea debris.22,23 Blight disease affects all parts of the shoot of chickpea plants, producing lesions, and shoot breakage.24 The disease significantly reduces chickpea seed yields and quality, and, depending on climatic conditions, yield losses for susceptible cultivars can reach 100 %.21 Economic losses due to blight damage have been substantial in many regions including Australia, Canada, Latin America, southern Europe, the United States of America, and West Asia.25
The use of biological control based on natural microorganisms offers a powerful alternative to synthetic chemical control of plant diseases. In fact, the abuse of chemical control agents, such as pesticides or fungicides, to cure or prevent plant diseases has often been reported to bring about a wide array of pernicious effects, particularly on plant, soil, environment, and, ultimately, human beings. The ten strains evaluated in this study were isolated from rhizosphere of healthy chickpea plants from four regions of West Azarbayjan. Rhizobacteria play an important role in identification of potential biocontrol agents, and that the strains should be from the rhizosphere of the target crop. Rhizospheric Pseudomonas strains were reported to be effective for the control of a wide range of fungal and bacterial diseases.26 Bacteria of the genus Pseudomonascomprise a large group of the active biocontrol strains as a result of their general ability to produce a diverse array of potent antifungal metabolites.
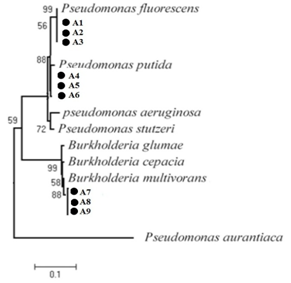
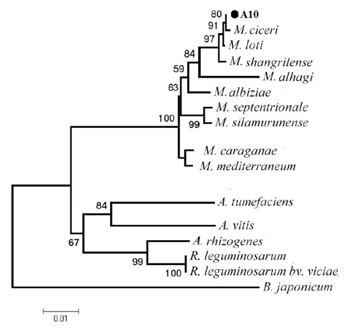
Molecular identification revealed that antagonistic strains belong to the species P. fluorescens (three strains), P. putida (three strains), B. multivorans (three strains) and M. ciceri (one strain). Antagonistic properties of strains tested in vitro were influenced by culture medium composition, the fungal pathogen, and its growth stages. P. fluorescens strains were found to be the most efficient by exhibiting the highest inhibition. Strains of P. fluorescens showed known biological control activity against certain soil-borne phytopathogenic fungi. In fact, Rajappan & Ramaraj27 evaluated the in vitro efficacy of P. fluorescens against the cauliflower wilt pathogen F. moniliforme. Also, Janisiewicz and Roitman28 have also reported that blue mold and grey mold of apples and pears could be controlled by Pseudomonas. Fluorescent Pseudomonasstrains were also found to be effective against Sclerotium rolfsii under greenhouse conditions in limiting groundnut and collar rot incidence. Enzymatic degradation of the cell wall of fungal pathogens by biocontrol agents has been reported.29 In this work, isolated bacteria have protease, cellulase, and chitinase activitiy.
Among the fungal diseases that affect chickpeas around the world, one of the most serious is the Ascochyta blight, caused by AR. Ascochyta blight is a highly destructive disease in most chickpea-growing areas of the world. The use of biological control based on natural microorganisms offers a powerful alternative to synthetic chemical control of plant diseases. In this study based on the phenotypic and sequencing of 16S rDNA and comparison with Gen Bank database of sequences the antagonistic strains were identified as species Pseudomonas fluorescens (three strains), P. putida (three strains), Bulkholderia multivorans(three strains) and Mezorhizobium ciceri(one strain). All the tested strains resulted in > 30% inhibition on PDA.
None.
The author declares no conflict of interest.

©2016 Azizpour, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.