Advances in
eISSN: 2373-6402


Research Article Volume 5 Issue 3
Division of Biochemistry, ICAR-Indian Agricultural Research Institute, India
Correspondence: Suresh Kumar, USDA Norman E Borlaug Fellow, IUSSTF Fellow, Principal Scientist,Division of Biochemistry, ICAR-Indian Agricultural Research Institute, New Delhi-110012, India, Tel +91-11-2584-2038, Fax 91-112-584-6420
Received: October 20, 2016 | Published: November 25, 2016
Citation: Kumar S, Singh A. Epigenetic regulation of abiotic stress tolerance in plants. Adv Plants Agric Res. 2016;5(3):517-521. DOI: 10.15406/apar.2016.05.00179
Sequence specific, genome-wide changes are being reported in plants which often correlate with regulation of gene expression at transcription levels. Many of such changes occur during stress exposure, and both gene expression and chromatin changes may revert to the pre-stress state shortly thereafter. There are reports for stress-induced chromatin changes that are transmitted to the progenies. Such changes alter gene expression without having any change in the DNA sequence. These epigenetic changes include modification in DNA base, histones and small non-coding RNAs. Some of the cytosine residues in nuclear genome may get methylated at 5’ position by the action of DNA methyl transferase. Histones are also subjected to post-translational modifications which affect transcription, replication, chromosome condensation/segregation, as well as DNA repair. Methylation of a promoter may repress gene transcription, while methylation of coding region of a gene may cause post-transcriptional gene silencing. Epigenetic changes may also be inherited over the generations in the form of epialleles, which are considered as a source of genetic variability for crop improvement. The roles of epigenetic changes in plant’s response to stress are becoming increasingly evident, suggesting that epigenetic mechanisms play important role in stress tolerance, acclimatization, adaptation and evolution processes. Therefore, it is worth investigating the epigenetic mechanisms of gene regulation in plants, and their possible exploitation in crop improvement.
Keywords: crop improvement, DNA methylation, evolution, gene regulation, histone modification, stress tolerance
Plants, being sessile in nature, are continuously exposed to environmental perturbations such as extreme temperatures, water supply, light intensity, non-optimal mineral composition in soil, pest/pathogen infestation and interaction with parasites or herbivores. Adverse environmental conditions distort plant’s growth, development and productivity. Hence, plants have evolved several strategies to cope with the environmental stresses by prompt and harmonized changes at transcriptional and post-transcriptional levels, including the epigenetic regulation of genes.1 Many traits of ecological/economic importance such as yield, drought tolerance and herbivore resistance, are complex in nature which are considered to be determined by the combined action of multiple genes. Plants use a range of sensing and signaling mechanisms to induce stress responses when challenged to the environmental stresses. Signaling includes the involvement of several phytohormones upon biotic and abiotic stresses.2 Nevertheless, there are growing evidences that chromatin modifications, siRNA and DNA methylation are involved in transcriptional and post-transcriptional control of gene expression, which are critical for stress responses.3 Recent studies indicate that heritable variations in economically important traits may also be caused by the underlying epigenetic variations.4 Epigenetic mechanisms have been implicated in the regulation of stress-associated genes.5 Epigenetics refers to the heritable changes in gene expression resulting from modifications of DNA and its associated chromatin proteins without changing the underlying nucleotide sequences.6 Epigenetic modifications may also be reversible, and it can be associated with inactivation as well as activation of genes.7 DNA methylation and histone modifications are influenced by various abiotic and biotic factors, resulting in plant adaptation.8 Among these epigenetic mechanisms, DNA methylation is currently the best understood. Further understanding the epigenetic mechanisms of gene regulation may not only provide basic information about the regulation of genes under the stress, but also a valuable platform for potential applications of genetic manipulation of plants towards enhanced tolerance to the environmental stresses.9 Thus, deciphering the epigenetic machineries towards tackling the environmental stresses in plants has become an important area of scientific investigations. Abiotic stresses have direct, negative effects on the biochemical and physiological processes associated with plant growth and development.10 Abiotic stresses may also cause heritable changes in DNA methylation leading towards the formation of epialleles,11 which may be defined as the genetically identical genes having distinct expression pattern due to the epigenetic changes. Heritable DNA methylation at specific loci due to environmental factor, which persists even in the absence of the triggering environmental factor, produces epiallele.12 Understanding the molecular mechanisms behind stress-induced epigenetic regulation of gene expression may facilitate breeding programs to improve plants which might further minimize the need for excessive genetic modification of crop plants.13 DNA methylation has been reported to play a key role in gene expression through RNA-directed DNA methylation (RdDM) of genes and by inducing histone modifications. Methylated cytosine (5-mC) has been reported to be involved in many important biological processes, including transposon movement, genome imprinting, and regulation of gene expression.14
DNA methylation: an important epigenetic mark
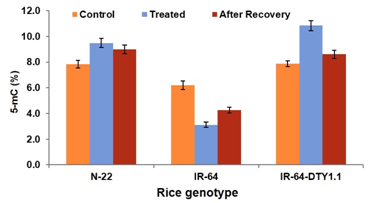
Four bases, adenine (A), cytosine (C), guanine (G) and thymine (T) are found in DNA, while T is replaced by uracil (U) in RNA. Methylated cytosine (5-mC, also known as the fifth base) has been found common in genomic DNA.15 Methyl-cytosine was identified long before the DNA was recognized as the genetic material.16 The content of 5-mC in genomic DNA varies considerably among the eukaryotes. In mammals, DNA methylation is restricted to the symmetric CG context, although non-CG methylation is prevalent in embryonic stem cells.17 However, nuclear genome of higher plants may contain more than 50% 5-mC in all the three different nucleotide sequence contexts: CG, CHG, and CHH (where H=C, T, or A).18 While CHG and CHH methylation is predominantly found in transposable elements (TEs), CG methylation is abundant in both TEs and genes.19 In both mammals and plants, centromeric and pericentromeric regions, as well as other repetitive elements are heavily methylated. In Arabidopsis, DNA methylation is highly concentrated in centromeric regions and repetitive sequences throughout the genome.20 The level and pattern of 5-mC are determined by DNA methylation and demethylation processes. Methylation can be removed from DNA by active, passive or both the mechanisms. For some genes, targeted methylation of C residues by methyltransferases may be sufficient to generate the desired methylation patterns, without the need of demethylases. For other genes, promiscuous methylation requires to be pruned by demethylases to create the desired methylation pattern. Demethylation may also be required to activate specific genes or to reset epigenetic state of the genome in response to environmental perturbations or during developmental stages.15 Stresses can induce changes in gene expression through hypomethylation or hypermethylation of DNA. In maize roots, cold stress-induced expression of ZmMI1 was correlated with a reduction in methylation in DNA of the nucleosome core. Even after seven days of recovery, cold-induced hypomethylation was not found to be restored to the basal level.21 We observed a considerable increase (20 and 37% in Nagina-22 and IR-64-DTY1.1, respectively) in global methylation of genomic DNA in the drought tolerant genotypes of rice under the stress. While the drought sensitive rice genotype (IR-64) showed decrease (46%) in cytosine methylation at genomic level on drought stress imposition (Figure 1). Moreover, 37% increase in global methylation was observed in IR-64-DTY1.1 merely because of the introgression of qDTY1.1 from Nagina-22 (showing only 20% increase in methylation) which could not be simply explained. Therefore, the role of the genetic background in the observed hypermethylation, if any, is being investigated. Even after ten days of recovery, drought-induced hypermethylation was not restored to the basal level (25 and 60% methylation was retained in N-22 and IR-64-DTY1.1, respectively), which might be involved in stress memory.
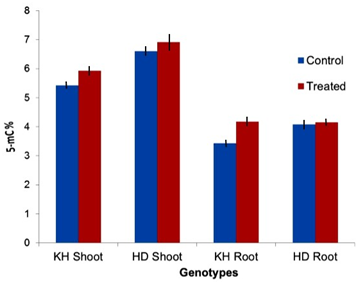
In wheat, we observed the basic methylation level to be higher (30-40%) in shoot tissues of the salt-sensitive (HD-2329) and salt-tolerant (Kharchia-65) genotypes compared to that of the root tissues. However, salt-induced increase in cytosine methylation at genomic level was found to be more in case of the salt-tolerant genotype (Figure 2). As roots experience the salt stress first, it must restrict entry of Na+ into the root cells. Increase in methylation in root tissues of the salt-tolerant genotype (Kharchia-65) might restrict the uptake of Na+ from the soil. No significant increase in methylation in the root tissue of HD-2329 could not limit the uptake of Na+ from the soil, which made it a salt-sensitive genotype. This indicated that the genes involved in salt tolerance might be regulated epigenetically and hypermethylation might down-regulate the genes, particularly the high-affinity potassium transporters (HKTs). Moreover, we observed differential expression of HKT genes in root and shoot tissues of the salt-tolerant and salt-sensitive bread wheat genotypes.1 Therefore, we targeted to investigate the role of cytosine methylation in regulation of expression of HKT1;4, HKT2;1 and HKT2;3 genes in the contrasting wheat genotypes. While we observed root-specific expression of the HKT1;4 gene, its expression was not observed to be regulated through DNA methylation. HKT2;1 and HKT2;3 genes were differentially expressed in both shoot and root of the genotypes. The salt stress-induced DNA demethylation in the coding region of the HKT2;1 and HKT2;3 genes was correlated with down-regulation of the HKT genes resulting into salt stress tolerance (our unpublished data)]. While most of the cytosine in CG context were methylated, the maximum variation in cytosine methylation was observed in the CHH context.
Histone modifications
Covalent modification of histone proteins is another primary mechanism of controlling gene expression. Histone modifications, particularly methylation of 9th lysine residue of histone H3 (H3-K9), cause chromatin condensation and block transcriptional initiations. Cytosine methylation can further reinforce histone modification patterns conducive to gene silencing.22 Some histone modifications such as acetylation, phosphorylation and ubiquitination have been reported to enhance gene transcription,23 while biotinylation and sumoylation have been reported to repress the gene expression.24 Trimethylation of H3K4 has been reported to activate transcription, while dimethylation of H3K9 and H3K27 was reported to repress transcription of the gene.23 In rice, submergence was reported to induce histone H3K4 trimethylation and H3 acetylation in alcohol dehydrogenase 1 and pyruvate decarboxylase 1 genes.25 These histone modifications were correlated with enhanced expression of the genes under the stress. However, the modifications were observed to be dynamic and were restored to the basal level after the stress was relieved. Stress-induced histone modifications can also influence DNA methylation. Knockout mutants and RNAi lines of stress-inducible histone deacetylase of Arabidopsisand maize showed increase in histone acetylation accompanied by changes in histone methylation pattern and derepression of silenced genes.26,27 Thus, dynamic histone modification marks could be converted into DNA methylation marks, which are often more stable.5
Micro-RNA (miRNA) and small interfering RNA (siRNA) are two important classes of small RNAs involved in regulation of gene expression. Genetic analysis of Arabidopsismutants impaired in genes for siRNA biogenesis/action revealed the involvement of siRNAs in RNA-directed DNA methylation (RdDM).28 Studies on repressor of silencing 1 (ros1) mutant of Arabidopsis revealed that DNA glycosylase ROS1 actively demethylates DNA by a base excision repair mechanism and can counteract RdDM.29 In Arabidopsis, a 24-nt siRNA was reported to down regulate the expression of P5CDH through mRNA cleavage, leading to decreased proline degradation, and enhanced proline accumulation and salt stress tolerance.30 Thus, stress-regulated siRNAs may also lead to changes in histone modifications and DNA methylation. Further studies are needed to unravel the roles of RdDM pathway in abiotic stress tolerance and their application in epigenetic manipulation for abiotic stress tolerance.
Epigenetic stress memory
Recent studies indicate that epigenetic mechanisms are indeed essential for stress memories and adaptation in plants. Ding et al.,31 reported that multiple exposures to drought stress enable plants to respond to a new stress by more rapid adaptive changes to gene expression patterns compared with the plants not previously exposed to the stress. They reported progressive accumulation (higher levels of transcripts than in the previous stress treatment) of RD29B and RAB18 transcripts in Arabidopsis. They suggested that the progressive change in gene expression and transcript accumulation may be the result of a progressive increase in H3K4me3 and phosphorylation of serine 5 (Ser5P) of RNA polymerase II during the process of recovery from the stress. They also observed that transcription level of the gene fallen down to the basal level present in unstressed plants, the higher levels of H3K4me3 and Ser5P in RNA polymerase II persisted, and may function as stress memory. Currently, we are studying biochemical and epigenetic changes in contrasting rice genotypes on multiple exposure to drought stress to understand the stress memory functions through the developmental stages as well as over the generation. Future studies in crop plants will undoubtedly provide insights into epigenetic stress memory and adaptation in plants.
Epigenetic variations and evolution
Heritable variations in plant phenotype, and thus potential for evolutionary changes, can in principle not only be caused by variation in DNA sequence, but also by the underlying epigenetic variations. However, the potential scope of such phenotypic effects and their evolutionary significance are largely unexplored. While working with epiRILs of Arabidopsis thaliana, Zhang et al.,32 provided evidence that variation in DNA methylation can cause substantial heritable variation in ecologically important plant traits, including root allocation, drought tolerance and nutrient plasticity, and that rapid evolution based on epigenetic variation alone may be possible. Thus, they demonstrated that variation in DNA methylation can cause heritability in ecologically important traits. An important next step, they suggested, to identify specific epigenomic regions underlying the variation. Paramutation, first described in maize, is a well-studied epigenetic phenomenon which is defined as an interaction between two alleles of a single locus resulting in a heritable change in one allele that is induced by the other allele.33 Recent studies demonstrated that MOP1, ZmRPD1, and RMR7 (orthologs of ArabidopsisRDR2, NRPD1, and NRPD2, respectively) are required for paramutation in maize, and suggested that RdDM pathway is needed for the establishment and maintenance of silencing in paramutation.34,35 A spontaneous mutant Epi-d1 in rice showed a metastable dwarf phenotype which is mitotically and meiotically heritable and corresponds to the metastable epigenetic silencing of the DWARF1 (D1) gene.36 The silenced state is correlated with DNA hypermethylation in the D1 promoter region. The epigenetic state of D1 is bidirectionally mutable, from active to repressed and from repressed to active. The bidirectional epigenetic state indicates that the epigenetic regulation of D1is due to de novo DNA methylation whichcould provide a mechanism of rapid adaptation to the changing environmental conditions.
Applications in crop improvement
Significant variations in epigenetic phenotypes in plants have been recognized over the last two decades. DNA methylation, a generator of epialleles, could have important implications for the plant breeders. Heritable epialleles are considered as a source of polymorphism and may have significant implications in plant breeding. Epialleles can emerge at higher frequency by far exceeding the rate of mutations that give rise to new alleles. However, their reversion rate is also high and emergence is affected by plant growth conditions. Data show that F1 hybrids are in general less methylated than their parental inbreds. The possible role of methylation in the expression of maize genes and performance of hybrids under different growth conditions has been examined in experiments with maize inbreds and hybrids.37 Repeated selfing for the isolation of inbreds, with emphasis on combining ability of inbreds, leads to gradual accumulation of methylated sites, which get released and/or repatterned when the inbreds are crossed to generate hybrid. Stressful growth conditions result in more methylated DNA, and such stress-induced methylation and suppression of genome activity could be at the core of higher yield of the hybrid. Manipulation of parental imprinting through epigenetic mechanisms may lead to the development of superior endosperm, which is a desirable trait for seed crops.38 Exploring the epigenetic mechanisms of seed development will eventually reveal the mysteries behind apomixis, i.e. an asexual mode of reproduction through seeds where embryos develop without meiosis and double fertilization leading to the production of fertile progenies identical to the mother plant.39,40 If this mechanism is applied successfully to the commercial crops, hybrid vigor can be maintained indefinitely, thus overcoming the current limitations of plant breeding to maintain hybrid vigor in more than one generation. Silencing of transgenes at both transcriptional and post-transcriptional levels is correlated with methylation of the transgenes. Methylation of the transgene promoter correlates with transcriptional gene silencing, while methylation of the coding region is associated with post-transcriptional gene silencing. Silencing of the transgenes has frequently been observed in plants, a major commercial risk, hampering the economic exploitation of transgenic plants. With the improved knowledge of the mechanism of epigenetically-imposed transgene silencing, there is tremendous commercial interest for avoiding it. The most efficient strategy suggested to avoid transgene silencing has been the careful designing of the transgene construct and thorough analysis of the transformants at molecular levels.41
There is growing evidences that epigenetic mechanisms contribute to stress responses and memory in plants. Therefore, it can be speculate that many of the observed trans generational stress responses could be due to epigenetic mechanisms. It has recently been shown that plants have an elaborate reprogramming system for epigenetic marks during sexual reproduction.42 It is possible that epigenetic machinery play more dynamic roles in crop plants, which have relatively higher populations of repetitive elements in gene-rich euchromatic regions. In fact, DNA methyl transferases and DNA demethylase mutations in rice showed more severe defects compared with the corresponding mutants in Arabidopsis.43-46 Gene manipulation for stress-tolerant transgenic plant development via DNA methylation initially needs a proper selection of the gene. Evaluating promoter sequence of the target gene for RdDM-associated features can be done using starPRO database. Subsequently, artificial siRNA from the targeted part of the gene can be introduced in the plants for stress tolerance. As host methylation machinery also plays important role to direct the DNA methylation, over-expression or silencing of methyl transferase genes and other key factors (e.g. siRNA biogenesis) can be combined; although these assumptions need validation. To understand the evolutionary dynamics and responses to ecological adaptations, mathematical models have recently been proposed which combine information on the probability of transmission of ancestral phenotypes, the number of epigenetic reset opportunities between generations, and assumptions on the environmental induction of epigenetically regulated traits.47 These models may facilitate identification of the heritable epigenetic variance and transmissibility for future molecular studies.
None.
The author declares no conflict of interest.

©2016 Kumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.