Advances in
eISSN: 2373-6402


Under conventional seed treatment, reports of efficacy loss or activity on non-target microorganisms foster the search for sustainable disease management alternatives. Biological seed treatment has been meeting these requirements and exerts benefits that go beyond plant health. Looking for those alternatives, 368 Endospore-forming bacterial strains were screened for the control of bacterial blight and damping-off, caused by Xanhomonas axonopodis pv. malvacearum and Colletotrichum gossypii var. cephalosporioides, respectively. From the sampled sites, the strains with the highest bacterial blight and damping-off control were obtained from Primavera do Leste and Campo Verde (MT), respectively. Those had the potential to control both diseases were obtained from Primavera do Leste. Consistent disease control with seed treatment was found with two strains: Bacillus subtilis UFLA285 and Paenibacillus lentimorbus MEN2 with disease symptoms reduced by 45 and 56%, respectively for damping-off and 26 and 76%, respectively for bacterial blight. Therefore beneficial bacteria are a plausible alternative for a sustainable disease control.
Keywords: bacterial blight, biocontrol, gossypium hirsutum, pgpr, bacillus, damping-off, eradication
Cgc, Colletotrichum gossypiivar cephalosporioides; Xam, Xanthomonas axonopodispv. malvacearum; AUDPC, area under the disease progress curve; HVS, highly virulent strain
The cotton diseases are limiting yield factors in the field, especially damping-off and ramulose caused by Colletotrichum gossypii var. cephalosporioides (Cgc).1 Another disease of apparently minor importance is bacterial blight caused by Xanthomonas axonopodis pv. malvacearum (Xam) which can be particularly harmful if a virulent race encounters the predominantly susceptible varieties.2 In order to achieve a successful yield increase, the amount of agrochemical active ingredients has increased over the last 10 years, placing the crop among the highest chemical consumer in Brazilian Agriculture.3
Fortunately, the used products has lead to a successful disease management over time4 accompanied by a cotton yield increase.5 Nevertheless, the amount of used ingredients may be reduced by successful eradication of pathogens such as C. gossypii var. cephalosporioides. The other major initiative in the cotton disease management program is the use of resistant cultivars. Presently, bacterial blight resistant cultivars have been adopted worldwide but, due to its unique agronomic features, growers insist on susceptible varieties such as Delta pine Acala 90 and DP90B, which increase the potential for disease outbreaks.2 Alternative disease control strategies such as biological control aims at reducing both the agrochemical use and selection pressure for resistant pathogen variants, assuring the sustainability of crop productivity.6
Biological control against foliar and soil-borne cotton diseases has achieved some level of success.7 Control levels of approximately 40% were achieved for bacterial blight by foliar treatments with several Bacillus isolates and damping-off by B. subtilis GB03, which is a commercial product that targets soil-borne diseases.
Because bacterial blight is a foliar disease and damping-off is soil/seed-borne, the selected biocontrol agents need to be specially adapted to survive and colonize these environments. UV light, temperature and humidity fluctuation associated with the leaf surface provide a less stable environment for bacterial growth than that encountered in the rhizosphere.8 Nevertheless, antagonists with broad-spectrum activity have been obtained by screening candidate microorganisms against multiple pathogens and by combining antagonistic bacteria to achieve synergistic effects,9 but to the best of our knowledge, there has not been reported a field efficacy of a biological control agent with broad spectrum activity in cotton. In this study, we aimed at isolating and selecting Endospore-forming bacteria with biological control activity against cotton bacterial blight and damping-off.
Sampling and isolation of endospore-forming bacteria
Samples from 200 sites representing the most important cotton-growing regions in Brazil were collected (Figure 1). At each sampling point, roots and soil loosely adhered to the roots (rhizospheric soil) were taken from the 5-10cm soil depth, placed into plastic bags, stored in ice or refrigerator (0-4°C) for no longer than two weeks, until isolation was done. Plants were sampled either because of their higher size and/or healthier aspect of their leaves as compared to neighboring plants. These characteristics were previously reported as possible effects of beneficial association with rhizobacteria in the field.10 Only sites continuously cropped to cotton for at least four years and containing up to 30-day old seedlings were sampled in order to improve the chances of obtaining isolates with good colonizing capacity and adaptation for growth on cotton root exudates.11
Isolation of endospore forming bacteria
Isolation of Endospore-forming bacteria for root endophytes and soil epiphytes were performed according to Bettiol.12 For rhizospheric soil epiphytes, 1g of the each sampled soil was added to a 15mL screw cap glass tube containing 9mL of autoclaved saline buffer (0.85% NaCl), vortexed for 1minute, incubated in an ultrasound water bath at room temperature for three minutes, the tube was then transferred to a water bath at 80°C for 10 m minutes, the sample was allowed to cool down at room temperature, vortexed again for three times and serially diluted by pipe ting 1mL to a new tube until 10-5. The last three dilutions were plated in nutrient agar, by pipe ting 0.1mL and spreading with a Drigalski’s loop. The plates were incubated in a growth cabinet at 28°C for 24h and the individual colonies for the last dilution for which bacterial growth could was obtained, the morphologically most frequent bacterium was purified in a new agar nutrient plate and stored at -80°C until use.
For root endophytes, each root was weighed, then washed in running tap water, and surface sterilized by transferring each root sample to a 70% ethanol solution for 30seconds, then to a 0.5% active chloride hypochlorite solution for 3minutes and washed two times in sterile distilled water. To confirm the efficiency of the surface sterilization, 0.1mL of the last washing water for each root sample was plated on nutrient agar. To isolate the endophytic bacteria, each root sample was transferred to a tube containing saline buffer in a volume enough to make up a 10 fold dilution according to each root fresh weight. The surface sterilized root was then crushed with a pestle, vortexed, incubated at 80°C, vorted, diluted, plated, purified and stored according to the described for the epiphytic bacterial isolates.
Characterizing endospore-forming strains
All obtained strains have been characterized as meeting the criterion of forming endospores. For that, each strain was grown for 48h in nutrient agar at 28°C and the bacterial growth was scratched from plate, eluted in 500mL of autoclaved tap water in a 1.5mL eppendorff tube, spread over a glass slide and processed for conventional gram staining to confirm the production of endospores.13 Strains not meeting those criteria have been discarded.
Preparation of artificially contaminated cotton seeds
Contaminated seeds were produced as described below to screen each biocontrol agent in the greenhouse for the treatment of Xam or Cgc inoculated seeds, using cv Deltapine Acala 90. Isolate Cgc1 of Cgc maintained at the Seed Pathology Laboratory (DFP, UFLA) was grown on PDA at 25ºC, with 12h light for eight days. A 200-μl aliquot of spore suspensions containing 105conidia/ml was spread on 9-cm Petri dishes containing PDA amended with mannitol (69.4g/l) to yield a -1MPa and seeds incubated for 3 days over a 5-day old fungal mat water potential.14
Seeds infected by Xam were obtained by vacuum infiltration. Isolate IB1153 maintained in the Bacteriology Laboratory (DFP, UFLA) was grown on medium 523,15 eluted to a 108cfu/ml cell suspension and transferred to a 500ml beaker, to which a 2ml aliquot of the bacterial cell suspension was added per gram of cotton seeds. The beaker was placed inside a desiccator, connected to an air pump that applied a 40cm lead (Hg) vacuum pressure for two minutes for two times. Non-inoculated control seeds were prepared by repeating the same procedure without the pathogen.
Screening bacterial isolates
A total of 208 endospore-forming isolates from rhizospheric soil and 153 from root endophytes were obtained in this study. Additionally, the bacterial isolate Paenibacillus lentimorbus MEN2 obtained from roots of Cucumis melo by Rosa Mariano was included in our screening16 as well as six other strains from our collection that were efficient in other experiments.
Experiments were conducted separately in a greenhouse for Cgc and Xam. All bacteria were individually grown on nutrient-agar medium for 48h at 25ºC and a suspension containing 108cells/ml was prepared in saline solution. Pathogen-infected cotton seeds were treated by immersion in the suspension (2ml/g seeds) for 30min, subsequently drained and used 12h later for the experiments. Negative controls were done by immersion of infected seeds into saline solution and positive controls by immersion of non-inoculated seeds into saline solution at the water rate used for the antagonist treatment. Five seeds were sown per pot containing 500 ml of a commercial potting mix (Eucatex®, Sao Paulo). Germination was evaluated 15 days after sowing and disease severity at germination and every three days up to 15days after sowing for both Cgc and Xam. Severity of Cgc was evaluated on the basis of a 0 to 3 disease index, adapted by Teixeira et al.17 and severity of Xam was evaluated using a 0 to 4 scale adapted by Ishida et al.18 Disease severity was transformed into disease index19 for each pot and used to calculate the area under the disease progress curve (AUDPC).20 Screening for each pathogen was split into five batches and the strains that resulted in the lowest AUDPC from each batch were tested again in a series of three experiments (biological replicates).
The strain that resulted in consistent disease control was fully identified by sequencing the 16SrRNA gene, using primers 8F (5’-AGAGTTTGATCATGG-3’) and 1492R (5’-TACCTTGTTACGACTT-3’), designed based on the Escherichia coli 16S rRNA NCBI deposited sequence, and following protocol previously described Barretti et al.21
Statistical analyses
All variables evaluated in the experiments were submitted to variance analysis after certifying normal distribution of the data using the SAS program. Tukey’s test at P≤0.05 was used to compare the means.
From all collected samples, endospore-forming bacteria were recovered in the rhizospheric soil but not always as endophyte. The soil population of endospore-forming bacteria was ca.100 times higher than the root one with lower variation for soil than for endophytic communities (Table1). For each sampled category, i.e. soil or root, the bacterial community was similar and averaged 3.93 and 2.01 log10cfu/g, respectively. Interestingly, the variability of root samples was much higher than the soil one.
The endophytic community of cotton is assumed to harbor log10 2-5 colonies of bacteria per gram of root tissue.22 The authors proposed that the variation in the bacterial community abundance was dependent on plant cultivar and age, the oldest the root, the highest the bacterial community. In our case, the bacterial community found in average varied very little among samples and was below the minimum bacterial community found by the authors. One has to consider that the plants have been sampled at early stages, which means that is expected for the bacterial community to be lower. Furthermore, in our isolation, we have only considered the endospore-forming bacteria and, even bacteria that would be able to form endospores but were still in the vegetative phase have not been recovered. Therefore the ca. log10 2 endospore forming bacteria may be considered a high number and may represent the importance of Firmicutes as an important group within the endophytic bacterial community.
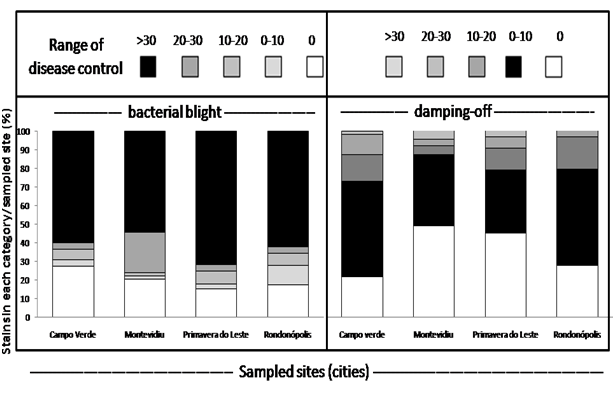
The efficacy of obtained strains to control bacterial blight and damping-off or both diseases was dependent on the sampled site (Table 2). Strains obtained from all sampled sites had a high percent of strains with more than 30% efficiency in bacterial blight control than in damping-off. The high endospore-forming bacterial community in cotton soils is a possible indicator of their potential in the higher suppressiveness to bacterial blight. Alvarado et al.,23 found that the endospore-forming bacterial community in the soil was the only soil attributes that significantly positively correlated with the suppressiveness and negatively with conduciveness of tropical soils to Pectobacterium carotovorum subsp. carotovorum.
In the screening of antagonists, there were differences in the efficacy of strains according to the location from where the strain was obtained (Table 2). For strains sampled from Primavera do Leste (MT), 70% of the obtained strains showed at least 30% efficiency on that disease control. Strains from Rondonopolis (MT) and Campo Verde (MT) had both 60% of strains with at least 30% bacterial blight control and Montevidiu was the site where 50% of the sampled strains assured such level of the disease control. From all sampled sites, the only location where the disease could be noticed on the cultivar Delta opal was in Montevidiu (Table 1) and this was the location where the lowest percent of isolated strains were effective in at least 30% for bacterial blight. In the field, for the management of the disease, grower use copper fungicides and this may have a direct impact on the reduction of beneficial bacterial communities, which may lead to more frequent disease epidemic out beak.24
Site location |
|
||||||||
|
Montevidiu |
Primavera do Leste |
Rondonopolis |
Campo Verde |
|||||
|
Soil |
Root |
Soil |
Root |
Soil |
Root |
Soil |
Root |
|
Number of Samples |
48 |
48 |
90 |
90 |
21 |
21 |
36 |
36 |
|
Total Endospore Former Community (Log10 Colony Forming unit/g ±Standard Error) |
3.91±0.04 |
2.18±0.15 |
3.73±0.006 |
2.28±0.12 |
4.00±0.00 |
1.06±0.14 |
4±0.00 |
1.94±0.13 |
|
Number of Obtained Endospore-Forming Isolates |
50 |
39 |
94 |
72 |
23 |
20 |
41 |
41 |
|
Remark/Comment |
Bacterial blight outbreaks were frequent |
Common losses due to soil-borne pathogens |
Seed producing farms with intensive crop rotation |
High losses due to root nematode |
|||||
Table 1 Clinical and biochemical variables of individuals with overweight-obesity
Number of strains in each class of disease control per location |
|||||
|
|
Campo verde |
Montevidiu |
Primavera do leste |
Rondonopolis |
Range of disease |
|
|
|
|
|
Bacterial blight |
0 |
25 |
20 |
30 |
9 |
0-10 |
2 |
1 |
3 |
3 |
|
10-20 |
3 |
1 |
8 |
1 |
|
20-30 |
2 |
18 |
5 |
1 |
|
>30% |
49 |
50 |
121 |
26 |
|
Damping-off |
0 |
16 |
45 |
66 |
11 |
0-10 |
41 |
39 |
50 |
24 |
|
10-20 |
12 |
3 |
25 |
6 |
|
20-30 |
8 |
2 |
17 |
2 |
|
>30% |
2 |
2 |
8 |
0 |
|
Table 2 Screening of endospore-forming antagonists for the control of bacterial blight (Xanthomonas malvacearum) and damping-off (Colletotrichum gossypii var. cephalosporoides)
SD, standard deviation; BMI, body mass index; WC, waist circumference; AC, abdominal circumference; HC, hip circumference; RER, respiratory exchange ratio; HR, hear rate.
When the screening for both diseases was considered, only 73 strains out of the 368 could assure a 30% control of bacterial blight and 10% control of damping-off. The experiment has been repeated twice with the same range of pre-selected strains and the only two strains that displayed disease control within this range at three consecutive screening was UFLA285 and MEN2 (Table 3).
Treatments1 |
Damping-Off Related Variables |
Bacterial Blight |
||
AUDPC2,5 |
Germination3,5(%) |
Dry Weight4,5 (g) |
AUBBPC |
|
Bacillus subtilis UFLA285 |
250.0 b |
80 a |
0.17 a |
29.3 b |
P. lentimorbusMEN2 |
187.5 c |
80 a |
0.22 a |
10.2 c |
Positive control |
- |
80 a |
0.17 a |
- |
Negative control |
458.3 a |
53 b |
0.06 b |
39.6 a |
Table 3 Control of two cotton (Gossypium hirsutum) seed-borne diseases: bacterial blight (Xanthomonas axonopodis pv. malvacearum and damping-off (caused by Colletotrichum gossypii var. cephalosporioides).Germination and shoot dry weight 15days after sowing through seed treatment with selected rhizobacteria: Bacillus subtilis UFLA285, Paenibacillus lentimorbus MEN2, non-inoculated or treated and inoculated untreated controls.
1treatments encompassed each selected bacterial strain (UFLA285 and MEN2), a positive control represented by non-inoculated followed by immersion in saline buffer (0.85% NaCl) and a negative control represented by inoculated seeds followed by immersion into saline buffer
2area under the disease progress curve (AUDPC), was calculated according to SHANER & FINNEY (1977)
3germination was calculated as the number of seedlings per pot with severity below 2 at the 15th day after sowing
4shoot dry weight of seedlings harvest 15days after sowing and oven dried at 70°C until constant weight
5means are average of three experiments that were statistically similar and were analyzed collectively; means followed by the same letter in the column are similar according to Tukey’s test (p≤0.05)
From this group of 73 strains 44% were from the Primavera do Leste site, 22% from Campo Verde, 20% from Montividiu and 14% from Rondonopolis. Noteworthy, at Primavera do Leste, the highest endophytic (log10 2.28 cfu) bacterial community was observed. In this region, cotton is grown for at least fifteen years and rarely any crop rotation or cover crop is used which may select exclusively for cotton symbiotic bacteria that can gain internal access to the plant. From this site was recovered the most promising strain, UFLA285 as a root endophyte (Tables 3). The identification of the most promising strain was based on 16S rRNA sequence and blasted to several Bacillus subtilis sequences with 97% homology.
Screening experiments
In the screening for damping-off control, the strains UFLA285 and MEN2 (Table 3) showed disease control. Since no difference was observed between the three biological replicates, data was analyzed collectively and is presented as average of replicates for all experiments per treatment. Both strains assured germination higher than inoculated control and this result (80%) was similar to the actual seed germination potential in the absence of the pathogen (non-inoculated control). The disease, measured by the area under the disease progress curve, was reduced by 59 and 45%, respectively by Paenibacillus lentimorbus MEN2, Bacillus subtilis UFLA285 and the shoot dry weight was similar to the non-inoculated control.
Although breeding lines have some degree of resistance to ramulose,25 they are not yet available to growers and the disease control relies on fungicide seed treatment and plant sprays throughout the plant cycle.1,4 In drastic seed sanitization by using sodium hypochloride, Soave26 observed a reduction in 45% of the disease, which was a result similar to the reduction mediated by UFLA285 treatment, an even higher reduction was obtained by treating seeds with MEN2 (59%) under greenhouse tests (Table 3).
The use of commercial chemical seed treatments result in high germination and low post-emergence damping-off, as recently confirmed by Chitarra et al.4 However, the germination for the untreated inoculated control found by those authors (89%) was much higher than the one obtained in our experiment (53%) (Table 3), which is an indication of the high inoculum pressure obtained by the water restriction method for Cgc inoculation.14 Nevertheless, the germination obtained by the biocontrol agents (MEN2 and UFLA285) was similar to the untreated non-inoculated control (80%).
Furthermore, either UFLA285 or MEN2 controlled bacterial blight (Table 3) by 26% and 74%, respectively. For the first time, the rhizobacterium-based biological control of Colletotrichum gossypii var. cephalosporioides-mediated damping-off or a simultaneous screening for control of both bacterial and fungal cotton pathogens has been addressed. In regard to bacterial blight, a seed treatment effective in the control of this disease has been reported either based on a biocontrol agent27 or fungicide,28 the last is the only integrated approach for cotton seed treatment, where a same strategy would be effective against both tested diseases. They found that tolyfluoanid, a fungicide already used in cotton treatment against C. gossypii was able to reduce the disease in up to 80%, but the application technology is based on overnight soaking of seeds in the fungicide suspension which is yet to be optimized for large scale use and/or not to interfere on seed early germination.
Although most grown cotton varieties are resistant to the predominant Xam races, a highly virulent strain (HVS) Huang et al.,11 has already been detected and only varieties carrying the B12 gene are resistant.2 Since this gene has not been introduced into commercial varieties, both selected bacterial strains, UFLA285 and MEN2, are important tools in the management of an eventual bacterial blight outbreak.
B. subtilisUFLA285 and P. lentimorbus MEN2 have broad spectrum disease control, acting on bacterial blight and damping-off in cotton.
The authors are thankful to all cotton growers that allowed sampling in their farms. Financial assistance was provided in part by a grant from the FAPEMIG (CAG APQ-2916-3.09/07). Scholarships were provided by CNPq to the all authors. The opinions expressed are those of the authors alone; trade names and commercial products are described in this article solely for the purpose of providing specific information and neither the authors nor our sponsors imply recommendation or endorsement of them.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.