Advances in
eISSN: 2573-2862


Long runs of homozygosity (ROHs) are frequently identified in cases interrogated by single nucleotide polymorphism (SNP) array. Presence of ROHs may be because of parental relatedness, chromosomal recombination or rearrangements and provides important clues regarding ancestral homozygosity, consanguinity or uniparental disomy. In this study we abort the use of SNP array in the detection of ROHs in an offspring of consanguineous parents and his siblings. All siblings had ROHs identified by SNP array on various chromosomes. One of them has an important ROH that harbor recessive mutation in BSDN gene. In summary, we have demonstrated that during the genetics evaluation of a patient affected by a rare disorder in the setting of consanguinity, SNP array analysis should be considered, except if the diagnosis is obvious.
Keywords: homozygosity, chromosomal recombination, identical alleles, DNA, hypercalciuria
Regions of homozygosity are more abundant in the human genome than previously thought. Homozygosis means that an individual has two copies of one chromosome or chromosome region but both were inherited from the same parent, i.e. these regions have two identical alleles.1 There are many terms to describe regions of homozygosity (eg, absence of heterozygosity, runs of homozygosity, loss of heterozygosity), and each one brings a lightly different meaning. In this work the term long runs of homozygosity (ROHs) was used to describe continuous chromosomes regions that have the same alleles and copy number state of 2. The minimal size of ROHs is generally set around 3 to 10Mb in clinical analyses and 0.5 to 1Mb in population genetic analyses.1,2 Many recessive genetic diseases are associated with parental consanguinity because individuals born into such families typically have ROHs. Detection of ROHs, in and of itself, is not diagnostic of any disease and may be clinically benign. On the other hand, all ROHs regions have the potential to port recessive mutations.1,3,4 Single nucleotide polymorphism (SNP) array method is a technology used to perform copy number analysis and to detect runs of homozygosity in all chromosomes simultaneously. SNP array method uses millions of probes (25-mer oligonucleotides) and has coverage for all chromosomes in high resolution. There are two types of probes: one used to assess genotypes (polymorphic probes) and another for assessing copy number (non-polymorphic probes). For copy number (CN) analysis, through fluorescent probes, patient DNA signal intensity is compared to signal intensity of a set of reference DNAs, indicating whether there is a gain or loss of genetic material (Figure 1A). For genotype analysis, polymorphic probes indicate which SNP is present in a particular chromosome region, the alleles are arbitrarily designated as A for one SNP and B for the other SNP and reveal which of the genotypes (eg, AB, BB or AA) is present along the entire chromosome (Figure 1B).5 To demonstrate the use of SNP array in ROHs detection, we present here a case of parental consanguinity that exemplifies this issue.
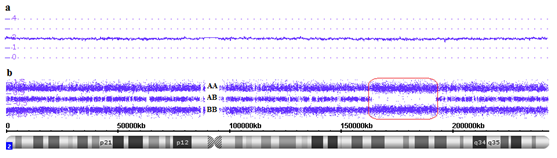
Figure 1 Affymetrix CytoScan® HD array showing chromosome 2. A. Smooth signal representing a normal copy number (purple line CN: 2.00) along the entire chromosome. B. Allele peaks representing AA, AB and BB alleles (three purple lines). Red box showing a long run of homozygosity (ROH), in this region there are only alleles AA or BB.
Patients
A deaf 20-year old male patient (S1) was admitted at the Hematology unit due to erythrocytosis detected before a knee operation. The investigation revealed normal leucocytes and platelet counts, high hemoglobin: 18.7g/dL, hypocellular bone marrow with an erythroid hyperplasia, normal erythropoietin: 19.2mUI/mL, normal ferritin, serum iron and transferrin, normal oxyhemoglobin dissociation curve (P50) and absence of mutations in the JAK2 (V617F and exon 12) and VHL genes. Additional laboratory tests revealed severe hypokalemia with potassium of 2.0mEq/L, bicarbonate: 23.0mmoL/L, normal creatinine: 0.9mg/dL, slightly low ionized calcium: 1.09mmoL/L, low phosphate: 2.2mg/dL, high parathyroid hormone: 120ng/L, slightly low Serum 25OH: 28.2ng/mL, high plasma aldosterone: 55.7ng/dL and renin: 65.0ng/mL, urinary volume of 3,530mL/day without hypercalciuria, very high urinary retinol-binding protein: 41mg/L. Normal blood pressure (110/70mmHg). Computed helical tomography excluded nephrocalcinosis. After hospital discharge, 200mg/day of spironolactone was added to the oral KCI supplementation for control of hypokalemia. The patient (S1) born premature after a complicated pregnancy by polyhydramnios, child of consanguineous parents. The patient had one deaf brother (S3), a stone-forming sister (S2) and another sister (S4) with no relevant clinical signs identified (Figure 2). The S1 case has already been published from the renal point of view.6 Here, we abort the use of SNP array in the detection of ROHs in an offspring of consanguineous parents and his siblings.
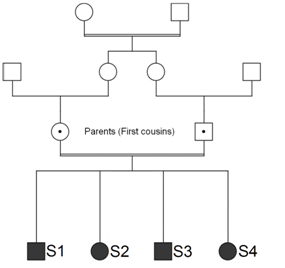
Figure 2 Pedigree siblings (S1, S2, S3 and S4) of consanguineous parents. All of them with long runs of homozygosity (ROHs).
SNP array analysis
SNP array analysis was performed for all peripheral blood samples (S1, S2, S3 and S4). Two hundred and fifty nanogram of DNA was extracted and digested, amplified, purified, fragmented, labeled and hybridized using Affymetrix CytoScan® HD Chip; according to the manufacturer’s instructions, Affymetrix GeneChip Scanner 3000 7G System was used for created CEL files. The Chromosome Analysis Suite v3.0 (ChAS) software was used for analyze CEL files.5 A minimal threshold for ROHs calls was 2 Mb.2
All siblings (S1, S2, S3 and S4) had ROHs identified by SNP array on various chromosomes (Figure 3). Detailed graphical results of all chromosomes can be seen in the Supplementary Figures (Chromosome1-Chromosome Y). When ROHs is found distributed throughout the genome, this observation is presumed to represent homozygosity caused by inheritance of genomic regions of regions identical by descent (IBD). When the parents of a proband share a recent common ancestor, their union is defined as consanguineous. The closer the parental relationship, the greater the proportion of shared alleles and, therefore, the greater the risk of the child (proband) inheriting 2 copies of a deleterious gene mutation, tumor suppressor gene or methylated genes from his parents.2 Although this has no immediate clinical utility and represents a pursuit largely of academic or social/ethical/legal interest, an estimate of the total proportion of the ROHs in the genome (represented by IBD) can be used as a rough assessment of degree of parental relationship. Here, S1, S2 and S3 had an IBD: 4,6% and S4 had an IBD: 3,1%. These data are consistent with third degree consanguinity, as illustrated in Table 1. We utilized fast online tool (Genomic Oligoarray and SNP array evaluation tool v1.0) that permits search genes in ROHs, associated with autosomal recessive disorders, using NCBI, UCSC and OMIM databases. Relevant OMIM Clinical Synopses can be searched, using key clinical terms permitting further filtering for disorders and candidate genes.7 Regions of ROHs present only in brothers with deafness (S1 and S3) were investigated in this online tool, which indicated five candidate recessive genes related to deafness: ORC1 (1p32), BSND (1p32.1), FREM2 (13q13.3), SMARCA4 (19p13.2), MAN2B1 (19p13.2).
Parental relationship |
Degree |
ROHs (IBD) predicted in child (~%) |
Parent/child |
First |
25 |
Full siblings |
First |
25 |
Half siblings |
Second |
12.5 |
Uncle/niece or aunt/nephew |
Second |
12.5 |
Double first cousins |
Second |
12.5 |
Grandparent/grandchild |
Second |
12.5 |
First cousins |
Third |
6 |
First cousins once removed |
Fourth |
3 |
Second cousins |
Fifth |
1.5 |
Third cousins |
Seventh |
<0.5 |
Table 1 Correlation between percentage of ROHs and degree of parental relationship
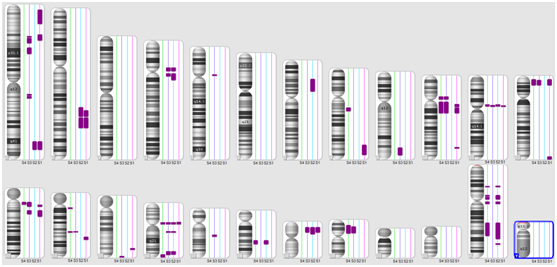
Figure 3 Karyoview (Affymetrix CytoScan® HD Array) showing multiple ROHs regions in S1, S2, S3 and S4 patients (purple bars next to each chromosome ideogram).
Despite the finding of low phosphatemia and high parathyroid hormone could have initially suggested the presence of some disorder of phosphate metabolism in the S1 case, the association of hearing impairment and hypokalemia led Nephrology unit to hypothesize about a late onset presentation of Bartter syndrome type IV. The diagnosis of S1 patient was confirmed by molecular analysis of the BSND gene that showed a homozygosity mutation (c.139G>A).The presence of erythrocitosis in a non-smoking person, in the absence of primary causes of polycythemia, is an intriguing feature. The cause of erythrocytes is investigated, including mutation search in the JAK2 and VHL genes, but the exact cause remains unclear. The BSND gene is located on chromosome 1. The brothers S1 and S3 have a ROH exactly in this gene location (Figure 4). The mutational analysis of patient S3 was not performed due to a lack of material and the impossibility of performing a new sample collection (The patient lives thousands of kilometers far from the medical facility). Bartter syndrome Type IV is a rare subtype of the Bartter syndromes that leads to both sensor neural deafness and severe renal salt wasting. This autosomal recessive disease is caused by mutations in the gene encoding barttin, an essential subunit of the ClC-K chloride channels expressed in renal and inner ear epithelia.8–10 This work report a case (S1) of a 20 years-old man with late onset presentation of Bartter syndrome Type IV and mild phenotype who had deafness, hypokalemia, erythrocytosis and secondary hyperparathyroidism. In summary, we have demonstrated that during the genetics evaluation of a patient affected by a rare disorder in the setting of consanguinity, SNP array analysis should be considered, except if the diagnosis is obvious.
Universidade Federal de Săo Paulo Ethics Committee (protocol CAAE 00547512.5.0000.5505).
We thank Mariana Cristina Lima Souza for providing JAK2 exon 12 and VHL analysis.
The SNP array test was supported by Research and Development from Fleury Group.
Author declares that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.