MOJ
eISSN: 2374-6920


Post-operative depression (POD) is a grave form of depression characterized by an increase in the intensity of the post-surgical acute pain, anxiety disorders, depression, bipolar disorder and personality disorders. Based on our work and published data, we will discuss a molecular hypothesis on the role of possible factors released by cancer tissue that could impact the Central Nervous System and lead to POD. Twelve proteins secreted by cancer tissue and transported by extracellular vesicles were identified as possible impacting factors on three neuronal metabolic pathways: the neuropentraxines/Cripto/AMPA system, the Neprilysin/Enkephaline metabolism and the Copper/Ceruloplasmin pathway. Our hypothesis posits that during disease progression a subtile balance in the metabolism regulation between cancer and brain is established and that this process could lead to the establishment of a mutual dependency. The algorithm is implemented as an R package and is distributed upon request.
Keywords: cancer, post-operative depression, bioinformatics, extracellular vesicles, secretome
POD, post-operative depression; MDD, major depressive disorder; EVs, extracellular vesicles; CNS, central nervous system
Cancer patients experience several kinds of emotional distress and mood disorders. Post-operative depression (or Post-Surgical Traumatic Stress Syndrome or POD for brevity) is a specific form of depression characterized by an increase of the intensity of the post-surgical acute pain1 and represents a peculiar condition that deserves to be investigated in more detail. Anxiety disorders, depression, bipolar disorder and personality disorders make up the plethora of patient mood alterations that may occur during the post-surgical period.2,3 Up to date it is not well known what factors cause POD and physicians often tend to associate it to a depressed state established during the period of disease experience. Several hypotheses are trying to connect POD to generalized causes of the post-operative condition such as anesthesia and disorientation.2,3 Here, we will try to discuss a molecular hypothesis, based on bioinformatic evidence, explaining how factors secreted by cancer influence the patient mood, not only during the development of the pathology but also after surgical tumor removal.
Molecules secreted by cancer tissue could be transported to the Central Nervous System (CNS) via Extracellular Vesicles (EVs). EVs comprise various vesicles that are released by cells into the extracellular space. Such vesicles include exosomes and smaller vesicles (microvesicles) that can contain and transport secreted proteins, metabolites and genetic material in form of RNA.4 EVs represent a device for intercellular complex communication networks in physiological metabolic conditions enabling long range organ-organ communication. EVs can carry out targeted deliveries of signals to cells with a specific cell surface receptor and protect information carrying molecules from various digestive enzymes in chronic diseases including various types of cancer. This mechanism may include signaling between cancer tissue and the CNS. Molecules secreted by cancer tissue and transported via EVs are generally ligands able to bind to receptors present in the CNS as well as in other organs. The analyses of microarrays of post-mortem brain biospecimens of patients affected by Major Depressive Disorder (MDD) surprisingly showed that many secreted ligands are poorly produced.5 This observation leads to the hypothesis that the depressed state of a cancer patient before the surgical tumor removal could predispose the CNS to receive the cancer tissue secreted ligands with more efficacy than in physiological conditions.
Software implementation and data preparation
To investigate connections between cancer and post-surgery depression, we wrote a program in R that creates a computational structure in which two distinct compartments represent two recipients equipped with two types of molecules: expressed receptors and secreted molecules. The structure can be described by the following expression:
BC[(R1...Rn)+(S1...Sn)];CC[(R1...Rn)+(S1...Sn)]
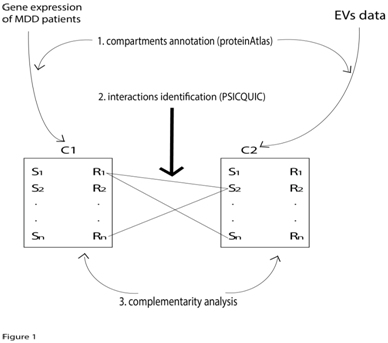
Here, BC represents the brain compartment; CC the cancer compartment, R and S represent receptor and secreted molecules, respectively. We called these two recipients "secretome" and "receptome" and they can be considered as transmitter devices in which the receptive component can be distinguished from the broadcaster component (Figure 1). As a broadcasting system, the two compartments can communicate with each other via serum/blood by means of EVs. The algorithm creates data repositories based on four distinct published databases:
Analysis workflow
In the following paragraphs, a schematic workflow of the analysis implemented by our algorithm will be presented. Results achieved by means of this pipeline will be discussed in the Results and Discussion section. The workflow is organized in three fundamental steps:
Compartment annotation: The MapSR function (MapSR stands for map Secretome and Receptome) annotates all the molecules expressed as receptors or secreted molecules of the two compartments. The Metamoodics gene expression dataset has been used for the brain compartment. In addition, MapSR generates annotations for the Cancer Compartment derived EVs using data from the EVpedia resource. The user calls the MapSR function with the omic parameter that specifies a user-defined –omics library created using the protein Atlas resource (i.e. secretome, or receptome). The user may specify the compartment to annotate as well as the p-value and fold change filter thresholds. The two libraries for the secretome and the receptome annotations are provided as internal repository included in the R code. For example, the code: CC_secretome <- MapSR (omic=secretome, compartment=CC, p value=0.001, fc=1.3, gt=TRUE, lt=FALSE)
BC_receptome <- MapSR (omic=receptome, compartment=BC, p value=0.001, fc=1.3, gt=TRUE, lt=FALSE)
creates two vectors, CC_secretome and BC_receptome, that contain the results of the MapSR function used for identifying and selecting all the molecules present in the Brain Compartment as receptors and in the Cancer Compartment as secreted proteins (filtered by p values=0.001 and fold change >=1.3).
Interaction identification: Once the two compartments are mapped, the user invokes a second function called FindInteractions that finds molecular interactions based on PSICQUIC8 among secreted proteins and their possible corresponding receptors. The function accepts three parameters in this form:
FindIteractions (PsicquicDataTable, CC_secretome, BC_receptome)
The first parameter refers to the PsicquicDataTable, another internal repository provided by the R code containing the PSICQUIC molecular interactions.8 The other two parameters are the names of the two compartments (vectors) created above.
Complementarity analysis: Exploiting the functionality of the embedded intersect function in R, it is possible to analyze the complementarity between compartments. Complementarity analysis aims at finding a set of secreted molecules present in one compartment that are down regulated in the other. For instance, to obtain a list of proteins secreted by cancer that could have an impact in the CNS of depressed patients and that are down regulated in these patients, we have to create the two compartments by calling the MapSR function as described in paragraph 5.2.1. Upon creation of the brain compartment, we apply filtering for down regulated proteins using the following call:
BC_down_sec <- MapSR (omic=secretome, compartment=BC, p value=0.001, fc=1.3, gt=FALSE, lt=TRUE)
Note that in this case we have set omic=secretome and the lower-than parameter (lt) to TRUE. By doing so, we select only those proteins that are down-regulated in the brain compartment. After the mapping we can intersect the two vectors with the intersect function:
Intersect (BC_down_sec, CC_secretome)
With this intersection we are able to identify possible molecular interlocutors expressed in cancer tissue that may have an impact on the CNS of the depressed patients (Figure 2) and Results and Discussion section.
Software availability: The algorithm is implemented as an R package and is available upon request.
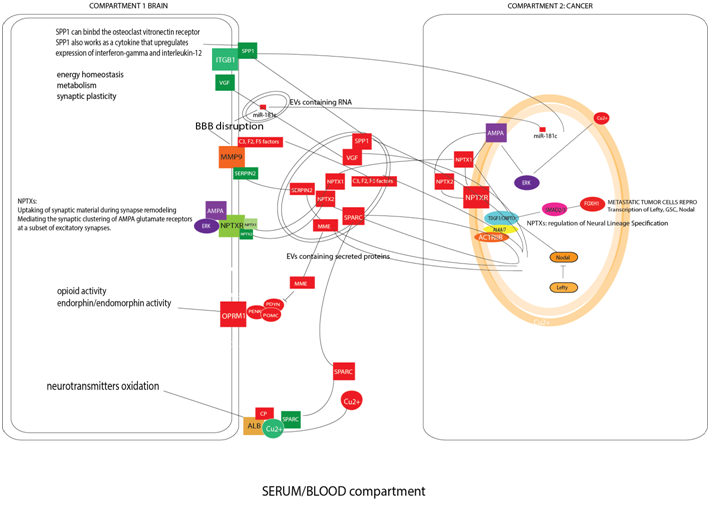
The algorithm selected 178 molecules that are down regulated in MDD patients and secreted in EVs (data not shown). Twelve of the 178 proteins were identified as possible impacting factors on neuronal metabolism according to the literature (Figure 1). In physiological conditions, the role of the Brain Blood Barrier (BBB) is to protect the CNS from the molecular injuries induced by cancer. For this reason, molecules released by cancer tissue should not pass through the barrier. However, our analysis revealed that one of the engaged pathways is composed of MMP9, SERPINE2, C3, F2, F5 coagulation factors and C1QB (complement component 1, q subcomponent, B chain). These factors may disrupt the BBB permitting the passage of other cancer derived molecules.9 Moreover, the microRNA miR-181c, released by cancer metastases via EVs, contributes to the disruption of the BBB.9
When the BBB is disrupted, factors released by cancer tissue enclosed in EVs could communicate with the CNS. We will now briefly describe three of the protein clusters released by cancer tissue in EVs that are involved in neuronal signaling mechanisms and that are summarized in Figure 1: the Neuropentraxines/Cripto/AMPA system, the Neprilysin/Enkephaline metabolism and the Copper/Ceruloplasmin pathway. Up to date, it is not clear how these pathways are connected to the monoamine neurotransmitters and neuromodulators metabolism and what their impact is in depression. A necessary precondition for having an impact on patient mood, however, is the ability of these molecules to cross the BBB, a step that may be facilitated by the release of the MMP9/SERPINE2/coagulation factor complex by cancer tissue. NPTX1, NPTX2 (two neuropentraxines, synaptic proteins related to C-reactive protein) are dramatically down-regulated in Major Depressive Disease subjects. Their receptor NPTXR is implied in neuronal plasticity and when it is detected in the cerebrospinal fluid it represents a potential biomarker for neurodegenerative diseases. NPTX1 and NPTX2 appear to be over-secreted by cancer tissue. NPTX1 is fundamental in the Epithelial-Mesenchymal Transition of melanoma metastasis acting on the Cripto receptor and promoting SMAD4 transcription and neural differentiation of the melanoma metastases.10,11 NPTX2 plays a role in clustering of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors at established synapses resulting in non-apoptotic cell death of dopaminergic nerve cells via inhibition of the ERK pathway.12 The same anti-apoptotic mechanism was identified in cancer, as demonstrated in studies of renal cell carcinoma expressing NPTX2.13
The second pathway regards the opioid system. The opioid system is formed by beta-endorphines such as POMC, PENK and PYDN that bind the opioid receptor OPRM1 that is expressed in the frontal cortex. It was documented that these molecules have anti-nociceptive effects, decreasing feelings of distress and that in depression disorders they are down regulated.14 The metalloproteases Neprilysin (MME) degrades a variety of bioactive peptides including beta-endorphines and is produced and released via EVs by bladder cancer, lung cancer, primary and metastatic colorectal cancer and renal cell carcinoma.6 The presence of the MME in blood/serum is in accordance with the diminution of the beta-endorphines in the depression correlated to cancer.
The third pathway is represented by the Ceruloplasmin (CP) and the Cu2+ metabolism. Cu2+ is important for neurotransmitter oxidation via the CP.15 In particular, the CP/Cu2+ system seems to be down regulated in MDD and CP is considered a marker of Major Depressive Disorder.15 In fact, neurotransmitters (serotonin, nor adrenaline, adrenaline and dopamine) are oxidized by CP with Cu2+ as a cofactor. Increased CP in the blood causes a decrease of Cu2+ concentration in serum and brain and this leads to the production of inappropriate enzymatic reactions in the neurotransmitter balance.15 CP is released by bladder, colorectal, ovarian, renal, lung cancer-derived exosomes.6 The molecules described here represent only a short list of the proteins secreted and released by cancer derived EVs. According to the literature, these molecules could be dramatically deregulated during the depressed state of the cancer patients prior to surgery. Other molecules such as SPARC and SPP1/Osteopontin are directly involved in regulating survival and neural plasticity by impacting neuronal metabolism, but they are also involved in melanoma aggressiveness.16 Moreover, the release of the neurosecretory protein VGF via EVs by colorectal cancer cells17 and its successive lack after cancer surgery could have a strong impact on the neuronal energy homeostasis of the patient (Figure 1).18
Our hypothesis, based on computational evidence, posits that during cancer progression a metabolic communication link between cancer and brain is established. Cancer tissue appears to be able to produce and release molecules needed to limit and contrast the depression related symptomatology of cancer patients. This communication link could explain the worsening of the mood status after surgery, leaving the patients with an aggravated form of post-operative depressive syndrome. POD, in turn, contributes to the worsening of the overall physical condition and may favor the development of metastases. In conclusion, Bioinformatics approaches may be used to single out new molecular liaisons between cancer and POD and contribute to elucidating new mechanisms of communication between cancer and the CNS. These observations may eventually suggest new supportive therapies.
We thank Prof. Valentina Brogna for her precious help, in the manuscript revision.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.