Journal of
eISSN: 2373-4469


GWAS, genome-wide association studies; CVD, cardiovascular disease; TNH-CS, toronto nutrigenomics and health cross-sectional; HC, hormonal contraceptives; LA, linoleic acid; TFA, trans fatty acids
Nutritional genomics or nutrigenomics is the study of how dietary molecules interact with the genetic architecture of a species governing the metabolic pathways contributing to the health of a population. The study of nutrigenomics contributes to our understanding of how diet and lifestyle effects the distribution of diseases under the influence of diet-genetic interactions. Nutrigenomic investigations combine’s insights generated from the meta-analysis of population-based studies, genome-wide association studies (GWAS), gene expression assays, lipidomics, metabolomics, metagenomics and proteomics to examine how the combination of genetic polymorphisms, diet, age, sex, ethnicity and lifestyle contribute to health and influence disease. Nutrigenomics at its core is a systems approach to the study of metabolic heterogeneity. Under certain conditions, the human genome displays a level of plasticity that is just now being understood. Recent advances in digital health that combines the power of big data analytics, cognitive computing and molecular diagnostics into an integrated platform have set the stage for a more comprehensive understanding of how to model the complex interactions influencing the genetic architectures contributing to human disease.
Nutrigenomics and health policy
Globally, national healthcare systems are faced with the growing challenge of designing interventions to address chronic diseases. In the U.S., the combination of cardiovascular disease, diabetes and obesity are creating are exerting a significant economic impact. According to the CDC, cardiovascular disease (CVD), diabetes, obesity, cancer and kidney disease accounts for more than 75% of the nation’s $2.7trillion in annual spending for medical care. With the state-of-the-art in biomedical research and the emergence of genomic approaches to understanding the nutrient-diet linkages, can nutrigenomics facilitate a systems approach to personalized nutrition and population health? The case can be made for applying a system approach to addressing chronic diseases using nutrigenomics.1 With advances in nutritional genomics, in the near future, dietitians and nutritionists will be able to provide personalized nutritional advice based on a combination of lifestyle factors and genetics. Globally, healthcare systems are faced with the growing challenge of demonstrating to stakeholders the value of adopting behaviors that contribute to personal health. The larger challenge is translating these behaviors into sustained dietary choices across heterogeneous groups of people with varying socio-economic status, sex, and lifestyle preferences. The first of the goals is for national public health systems to direct the outputs of nutrigenomic research into policies that reduce the risks of disease at the population level. A second goal is to integrate nutrigenomics outputs into nutritional value chains and incentivize the development of food innovations that enable personal nutrition. Third, collaborate with stakeholders to ensure the quality and integrity of health communications translating the value of nutrigenomic information to individuals making choices to support health and wellness.
Nutrigenomic biomarkers, fatty acid metabolism and cardiovascular disease across populations
A considerable amount of progress has been made in understanding the linkage between lifestyle and CVD. Looking at the larger challenge of translating nutrigenomics across heterogeneous populations. How can nutrigenomics enable the adoption of personal nutrition approaches? The Toronto Nutrigenomics and Health Cross-Sectional (TNH-CS) study2,3 is an example of a groundbreaking cross-sectional study examining the combination of ethnicity, sex, lifestyle and genetic variation on fatty acid metabolism. Investigators measured the plasma fatty acid concentrations in two population-based cohorts of young Canadian adults (786 subjects) Caucasian (n=411) and East Asian (n=375). The investigation explored the combinatory effect of factors including sex, ethnicity, hormonal contraceptives (HC) and the genetic polymorphisms within the Fatty acid desaturase genes (FADS1 and FADS2) that encode the enzymes delta-5-desaturase (D5D) and delta-6-desaturase (D6D). 19 single nucleotide polymorphisms (SNPs) were examined, and several SNPs (9 in Caucasians and 8 in Asians) were associated with various desaturase activities. D5D and D6D are essential enzymes in the conversion of n=6 and n-3 polyunsaturated fatty acids (PUFA) to their longer chain products. Through a series of desaturation and elongation events, the n-6 PUFA linoleic acid (LA) is converted to arachidonic acid (AA) while the n-3 PUFA α-linolenic acid (ALA) is converted to eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA).
Several factors have been implicated in the control of endogenous fatty acid levels. These include diet, genetic variations and hormonal regulation. Single nucleotide polymorphisms (SNP) in the FADS gene cluster have been shown to be associated with increased Homeostasis Model Assessment of insulin resistance (HOMA-IR), as well as High-sensitivity C-reactive protein (hsCRP) levels, both of which are indicators associated with abdominal obesity, metabolic syndrome and atherosclerosis. FADS polymorphisms have been shown to explain 28% of variability in AA levels in serum phospholipid fatty acids in Caucasian participants. AA is a primary precursor for the synthesis of pro-inflammatory eicosanoids as well as the vasoconstrictor TXB2, and AA generated prostaglandins which are shown to be pro-arrhythmic. This also suggests that associations between FADS polymorphism and altered n-6 metabolism in Caucasian females may contribute to a modified risk of inflammatory and cardiovascular conditions through the alteration of cardiovascular concentrations of AA. The THN-CS study identified sex and ethnic-specific effects of FADS polymorphisms on desaturase activity. Unpredictably, the investigation also determined that the genetic polymorphisms examined have less influence in the conversion of EPA to DHA in Caucasian men, East Asian men and East Asian women. However, FADS SNPs when combined with HC use played a prominent role in this pathway, specifically in Caucasian women. These findings highlight the differential roles of sex, ethnicity, and FADS polymorphisms within specific portions of the fatty acid metabolic pathway.
The THN-CS study is significant in that it provides us with an insight into the complexity faced by public health systems in designing interventions for heterogeneous populations. For population health advocates the challenge of who is at risk needs to be addressed. The observation that the metabolic outputs of the FADS gene cluster differs across populations provides us with an insight into how sex and lifestyle combine to influence metabolism and predispose people to the risk of CVD. Can using a nutrigenomics approaches provide us with the resolution necessary to differentiate SNPs associated with a higher risk of CVD from those SNPs with a lower risk, using the FADS-fatty acid diet-gene interaction model? A study conducted at the Wuhan Asian Heart Hospital (WAHH) explored the association of FADS gene SNPs with plasma fatty acids in Chinese Han patients with coronary artery disease (CAD).4 5 SNPs (chromosome 11q12-13) showing the greatest variability in the FADS gene cluster (FADS1, FADS2, and FADS3) were selected for evaluation. High-resolution melting was used to identify a SNP rs174460 C allele that is associated with a higher risk of CAD. It also identified a SNP rs174537 T allele that is associated with a lower risk of CAD. In this study, the fatty acid levels of genetically (n=1015) unrelated Chinese subjects (33-85 years) were evaluated (n=505 CAD, n=510 controls). The study found that 3 desaturase activities (D5D, D6D, and D9D) were associated with CAD in a Chinese Han population. Compared to controls, the plasma fatty acid composition in CAD patients were significantly different. CAD patients also had lower levels of the essential fatty acid LA than the control group. This is significant because LA is considered to be a major factor influencing arterial stiffness.
FADS biomarkers and fatty acids composition interact to influence cardiovascular health and metabolism and contribute to health disparities
Recent reports highlighting the influence of fatty acid composition on metabolic syndrome and arterial stiffness, such as LA and dihomo-α-linolenic acid (DGLA). The investigators hypothesized that sufficient concentrations of LA in the diet could potentially improve insulin sensitivity and lower the risk of coronary heart disease. Higher plasma concentrations of LA have also been associated with lower inflammation and lower cardiovascular risk. However, this study demonstrates that carriers of the rs174460 C allele (FADS3 gene) had higher levels of DGLA. This is significant because DGLA is endogenously synthesized and it is not linked to dietary intake. However, its presence may indicate impaired fatty acid metabolism and contribute to long-term metabolic disorders. For carriers of the rs174537 T allele (FADS1 gene) their lower risk of CAD was associated with their reduced capacity to convert oleic acid to stearic acid. The SNP rs174537 accounts for up to 19% of the variation in AA levels. However, for carriers of the SNP rs174537 G allele, higher circulating levels of plasma AA have been observed. In a recent study, tests for association in individuals across 80 SNPs in the FADS gene cluster, determined that African Americans have significantly higher circulating levels of plasma AA than European Americans. It was determined that 79-82% of African American carry two copies of the G allele compared to 42-45% of European Americans. The allelic effect of the G allele, which is associated with enhanced conversion of DGLA to AA, on enzymatic efficiency was similar in both groups.5
Parallels can be drawn between the risks attributed to the higher AA levels observed in Caucasian females in the THN-CS study, and health disparities observed in the African American population that continues to experience higher incidence of in CVD. The results of the WAHH study are significant in that they demonstrate the capacity to use nutrigenomics to differentiate SNPs of physiological importance to CVD within a genetically distinct population of Han Chinese ancestry. Furthermore, this study provides some interesting hypothesis about the potential consequences of fatty acid composition on disease. The implications of these findings are exciting in that food innovators could use an understanding of the distribution of SNPs to supply mixtures of dietary lipids to improve insulin sensitivity and lower the risk of CVD in carriers of the rs174660 SNP. Conversely, based upon their allelic distribution (G or C) carriers of the rs174537 SNP could be directed to select products with nutrient-fatty acid combinations that support cardiovascular health, cognition and immunity.6 An interesting set of questions arises from this review: First, are the tools available to practice personal nutrition and leverage insights gained from nutrigenomics to reduce health risks? At this time, we do not have a comprehensive set of tools to address every foreseeable challenge. But with the tools that we do have, we can genotype an individual’s FADS genes and look for SNPs associated with the risk of CVD. Considerable product development must continue to integrate mobile systems that enable health information consumers to create value in their daily lives. In addition to genotyping, proteomic analysis, and metabolomic analysis may be recommended depending on the client’s health status.
Second, in an environment where people are chronically exposed to trans fatty acids (TFA), can we identify the populations that are most at risk of CVD? TFA is found in fried foods, commercial baked goods and processed foods, and have been linked to adverse health outcomes. Considering the fact that men and women can differentially respond to TFAs depending upon their allelic variation of the FADS SNPs, lifestyle and racial population.7 The results from the TFA-CHARGE study has established that we can begin to build and test a prototype system that integrates SNP data with personal information, pharmacology and toxicology studies to understand who is at risk. Briefly, investigators in the TFA-CHARGE study performed a meta-analyses using 7 cohorts of European-ancestry (n=8013). Using data obtained from GWAS they were able to evaluate genome-wide variation in SNPs associated with circulating TFA. SNPs with genome-wide significant associations were also identified among African Americans (n=1082), Chinese Americans (n=669), and Hispanic Americans (n=657). The SNP analysis identified a single SNP rs174548 near the FADS gene cluster that was associated with cis/trans-18:2 for members of the European-ancestry, the Hispanic Americans, and Chinese Americans cohorts but not the African Americans. A second SNP rs174579 in FADS2 was discovered in the African American cohort that associated with cis/trans-18:2. Further investigation revealed that in the presence of circulating n-3 and n-6 PUFA, the association between SNP rs174548 and cis/trans-18:2 remained significant, except when in the presence of arachidonic acid (AA). Later it was demonstrated that in the presence of AA, the association between rs174548 and cis/trans-18:2 could be significantly reduced in the European-ancestry participants, with similar reductions in Hispanic Americans and Chinese Americans.8
In terms of predicting who is at risk, these findings demonstrate how TFAs in the diet interact with the genetic architecture of the FADS gene cluster to affect different outcomes according to the population’s racial-ethnic identify. However, the risk of CVD is not solely attributed to polymorphisms in the FADS gene cluster. In order for our prototype to display the fidelity necessary to predict which combinations of factors present the greatest risk and aid nutritionists in providing the health information that consumers require, we need to incorporate an understanding of how multi-gene networks contribution to the spectrum of cardiovascular diseases. Investigators in the DNL-CHARGE study, identified polymorphisms in 7 novel loci associated with circulating levels of saturated fatty acids: palmitic acid(16:0), stearic acid(18:0), palmitoleic acid(16:1n-7), and oleic acid(18:1n-9). These fatty acids are synthesized via de novo lipogenesis (DNL) and are the main saturated and mono-unsaturated fatty acids in the diet. Levels of these fatty acids have been linked to diseases including type 2 diabetes and coronary heart disease.9 The DNL-CHARGE study examined 8,961 subjects of European-ancestry in 5 cohort studies: 2 cohort studies in the Cohort for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium: the Atherosclerosis Risk in Communities (ARIC) Study and the Cardiovascular Health Study (CHS). Data were also obtained from 3 additional cohorts: the Coronary Artery Risk Development in Young Adults (CARDIA), the Invecchiare in Chianti (InCHIANTI) Study, and the Multi-Ethnic Study of Atherosclerosis (MESA). Only subjects of European ancestry from each cohort were included in their analysis.
The 7 loci associated with plasma fatty acids are presented with a description of the protein:10
A systems approach: its promise to reduce health risks and stimulate health innovation
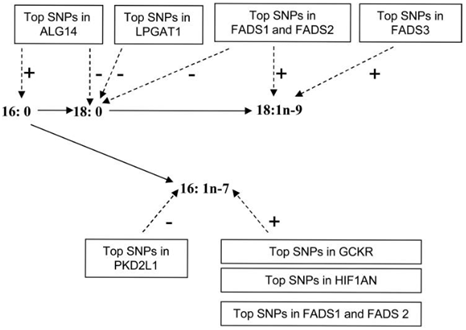
Figure 1 Summary of genome-wide association results associated with plasma fatty acids. The genome-wide significant associations of identified loci (and genes of potential interest) with each fatty acid are shown with dashed arrows and the +/- signs indicate the direction of the associations for the minor alleles at each loci.9
Healthcare consumers recognize that the key to reducing their risks of cardiovascular disease and long-term metabolic disorders are multifaceted. In 2013, the market size of the healthy eating, nutrition and weight loss segment of the global wellness industry was estimated to be $574billion dollars.11 In short, consumers are looking for innovative products and services to support their health and wellness and reduce their risks of disease. Healthcare consumers want to know what impact genetic variation has on their personal health. Patient advocacy groups, public health and healthcare systems administrators want to understand the spectrum of disease present within a given population and quantify the risks of predisposition associated with lifestyle factors and health disparities. Recently, GWAS in patients with coronary artery disease revealed about 30 genetic loci that are considered to represent the most significant genetic-disease susceptibility factors. Several of these genes had been previously identified, but the majority have not been associated with any known risk factor or signaling pathway that contributes to the development of atherosclerosis. A systems approach has been proposed to gain a deeper understanding of how genetic variation within multi-gene networks contribute to heart disease.12 By analogy, a systems approach can also be applied to understanding the complex interactions between various classes of nutritional molecules, metabolites, and the genetic loci comprising the biological networks that contribute to the spectrum of disorders observed across different human populations that are influenced by diet and lifestyle.
The gut microbiome functional foods and cardiometabolic disease
The findings from the DNL-CHARGE study opens up a new set of possibilities where a systems approach can be used to inform health innovation. Equipped with an understanding of the biological networks that contribute to diet-influenced outcomes, innovative products can be designed to enable individuals and populations to benefit from nutritional interventions designed to address cardiometabolic diseases. Mounting evidence supports the hypothesis that functional foods containing physiologically-active components may be healthful.13 Recent human and animal studies indicate clinical and mechanistic links between heart disease and intestinal microbial metabolism of certain dietary nutrients (carnitine, choline and phosphatidylcholine) producing trimethyl-amine N-oxide (TMAO). TMAO is a microbiota-dependent metabolite that participates in atherosclerotic plaque formation.14 In a set of experiments obtaining cecal microbial contents from two strains of mice: an atherosclerosis-prone and high TMAO producing C57BL/6J strain, and an atherosclerosis-resistant low TMAO producing NZW/LacJ strain. Cecal microbial contents from these donors were transplanted into apolipoprotein e null mice in which resident intestinal microbes were first suppressed with antibiotics. The results demonstrated that the mice receiving the C57BL/6J microbial transplants when fed a diet high in choline exhibited transiently high levels in TMA/TMAO. These mice also demonstrated choline-dependent enhancement of atherosclerotic plaque formation as compared with the mice receiving the NZW/LacJ cecal microbes. This was the first demonstration that atherosclerosis susceptibility can be transmitted via transplantation of gut microbiota. The study recognizes that gut microbes represent a novel therapeutic target for modulating atherosclerosis susceptibility.14 Recently, at the 11th International Functional Foods Workshop data was presented that supports continued investments into the development of functional foods that impact cardiovascular health.15 Metagenomic studies of 12 patients examining the link between atherosclerosis and gut microbiota determined that the guts of patients symptomatic for atherosclerosis were enriched in the genus Collinsella whereas controls were enriched in Eubacterium and Roseburia. At the functional level, patient metagenomes were enriched in genes encoding peptidoglycan biosynthesis whereas those of healthy controls were enriched in phytoene dehydrogenase genes. The finding of enriched levels of phytoene dehydrogenase was associated with elevated levels of β-carotene in serum. The possible production of this anti-oxidant by the gut microbiota may have a positive health benefit.16,17 From a systems perspective, the novel approaches of metagenomics and metaproteomics provide data that allow the detection of patterns under different physiological states, such as different diets, disease condition and antibiotic therapy. These large-scale patterns can be correlated with certain health or disease conditions and support the identification of novel therapeutic targets. Human beings have had the knowledge and expertise to cultivate probiotic products like yogurt for thousands of years.
This review considers how recent advances in nutrigenomics can be applied to understanding cardiometabolic disease. Several questions were asked to frame the examination and gain insight into the current state of understanding. In answer to the question, are the tools available to practice personal nutrition and leverage insights gained from nutrigenomics to reduce health risks? The answer is it depends upon the outcome you are trying to accomplish. Next-generation sequencing tools provides the ability to characterize genomic biomarkers involved in metabolism. Metagenomic methods allow for the characterization of patient microbiomes and compare the biosynthetic pathways contributing to their metabolic phenotype. As more genomic data is accumulated on increasing numbers of people, greater resolution will provide the support for increased utilization across populations and public health policy decisions. Iceland’s genomic health initiative and the establishment of deCODE genetics are examples of efforts to identify genetic variations associated with human disease. Because of the tremendous heterogeneity of the US population and the challenges to address health disparities and cardiometabolic disease across these populations, efforts to apply the science and support a learning healthcare system could begin to produce benefits to individuals, and inform members of various ethnic groups about the advantages of nutritional approaches for health and wellness. Healthcare consumers recognize that the key to reducing their risks of cardiovascular disease and long-term metabolic disorders are multifaceted. In 2013, the market size of the healthy eating, nutrition and weight loss segment of the global wellness industry was estimated to be $574billion dollars. sA second unanswered question remains. In an environment where people are chronically exposed to trans fatty acids can we identify the populations that are most at risk of cardiovascular disease? Considering the evidence examined in this review the answer is we should begin to identify those most at risk with the tools available at this time. According to a recent report commissioned by the American Heart Association, costs associated with heart disease in the U.S. will reach $818.1billion a year by 2030. In an attempt to make the case for utilization sooner than later, let’s examine the data from the Early Childhood Longitudinal Study, Kindergarten Class of 1998-1999. This study examined the incidence of obesity in 7738 participants and found that a third of children who were overweight in kindergarten were obese by eighth grade. And almost every child who was very obese remained that way. The both of these reports highlight the opportunity to leverage the potential of nutrigenomics to generate the data necessary to support a learning healthcare system and make data-driven decisions to improve the health of the country.
None.
Author declares that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.