Journal of
eISSN: 2373-4469


Harsh environmental conditions liked rought and salinity limit agricultural systems by affecting plant growth and production during critical developmental stages. The genetic improvement of agronomic crops to biotic and abiotic stresses is critical to ensure world food security; however many agronomic crops lack the genetic ability to tolerate stress demanding new genetic resources. This review highlights new strategies to improve tolerance to abiotic stress through the discovery and utilization of new genetic repertoires from plants grown in natural and desert ecosystems. Understanding the molecular basis of tolerance to abiotic stress will facilitate the development of innovative techniques to improve the performance of agronomic crops to stress. Furthermore, metagenomics can be used to examine how plants tolerate abiotic stress by characterizing the functional roles of microbial communities associated with plants growing in adverse environmental conditions. Altogether, the implementation of new genomic approaches is necessary to increase agronomic crop cultivation and overcome problems caused by abiotic stress.
Keywords:abiotic stress, genomic improvement, metagenomics, microbiomes
ABA, abscisic acid; SnRK, snf1-related protein kinase; LEA, late embryogenesis–abundant; DRE, dehydration-responsive element; CRT, c-repeat; ros, reactive oxygen species; RNA-Seq, rna sequencing; PGPB, plant growth-promoting bacteria; GIS, geographical information system; GPS, global positioning system
The world population is expected to reach about 8.3billion by 2030 with an estimated increase of 22% from the current population, which will put considerable pressure on food security as demand increases. In addition to biotic stress, another important factor that significantly reduces yield and limits cultivation land area is abiotic stress such as drought and salinity. For example, water stress can reduce average yield more than 50%.1 Additionally, climate change will further reduce the water availability and cause more droughts in the future. Drought caused by water deficit, extreme temperatures and low atmospheric humidity limits plant performance by affecting the level of endogenous phytohormones such as ABA and ethylene.2,3 Drought inhibits plant growth, causes osmolyte accumulation in plants, increases antioxidant enzyme activities and reduces yield.3 Water stress also reduces the rate of photosynthesis, suppresses chlorophyll synthesis and decreases the activity of photosynthetic enzymes,4 in addition to reducing cell division and end- reduplication.5 In general, plants respond to salinity and drought stresses that cause dehydration and iron imbalance with shared signaling pathways.
Abscisic acid (ABA) signaling pathway is an important hormone pathway in plant response to abiotic stress. ABA-deficient mutants are more sensitive to stress compared to wild type plants indicating ABA may play a crucial role during abiotic stress response. A recent work6 demonstrated increased levels of ABA in drought tolerant soybean cultivars grown under drought conditions compared with drought sensitive cultivars. The SNF1-related protein kinase (SnRK) family involved in ABA signaling transduction and transcriptional activation of multiple genes7 enhances tolerance to various stresses including drought, hyperosmotic conditions, cold and salt.8 For example, overexpression of a wheat TaSnRK2.4 in Arabidopsis thaliana enhanced the growth of primary roots and increased their tolerance to cold, drought and salt stress.8 Altogether, the Ca2+ and ABA signaling pathways mediate the expression of the late embryogenesis–abundant (LEA)-type genes including the dehydration-responsive element (DRE)/C-repeat (CRT), a class of stress-responsive genes that reflects damage repair pathways. In genomic studies of major crops grown under drought stress, transcriptomic and proteomic screens have demonstrated the importance of reactive oxygen species (ROS) scavenging, cell signaling, cell wall modification and phytohormone response.9 Transgenic lines have been used to evaluate roots grown under low water stress conditions. For example, overexpression of a metallothionein gene, OsMT1a, in rice improved growth under osmotic and dehydration stress conditions compared to control plants.10 Additionally, overexpression of NAC transcription factors led to increased tolerance to drought, salt and/or other abiotic stresses such as freezing tolerance.11 Due to narrow genetic backgrounds, many agronomic crops are still sensitive to abiotic stress demanding the use of new genomic strategies to improve plant tolerance to abiotic stress, which will ultimately increase food supply.
Arid and semi-arid regions represent about 40% of the world’s land area. This also includes the declining acreage of arable land in many parts of the world affected by limited irrigation. Furthermore, altered precipitation patterns onset by climate change will increase the acreage of the dry land. Therefore, this research is essential to ensure world food security; however many agronomic crops lack the genetic ability to tolerate stress demanding the genetic improvement of agronomic crops and the exploitation of new genetic repertoires. One important task to improve the tolerance of agronomic crops to abiotic stress is to explore genetic diversity found in natural and desert ecosystems. The ability of desert plants to grow under abiotic stress makes them relevant genetic resources for drought tolerance genes. For example, mangrove, (Avicennia marina) is a halophyte plant that can survive under high salinity conditions up to 600 mMNaCl and secrete salt with a lower crystal density. The high salt tolerance of A. marina is a consequence of its water use efficiency, which balances carbon gain, water loss and ion uptake by keeping the transpiration stream of these molecules at a low but constant level. Inorganic ion uptake is regulated partially by exclusion at the roots and via salt glands allowing A. marina to grow in coastal regions with up to a 9% salt concentration. In addition to the morphological and physiological traits associated with the ability of mangroves to survive under harsh conditions, tolerance to extreme environmental conditions is also attributed to the regulation of gene networks. This halophytic species is an ideal target for understanding molecular mechanisms underlying its ecological adaptation to high salinity and exploring genes for further genetic improvement of agronomic crops. Despite the ecological effectiveness of mangrove, understanding its molecular mechanisms to survive in such conditions remains elusive. One main limitation of genomic research on mangroves, is a lack of genomic and transcriptomic resources. More research projects are needed to employ new technologies like RNA-Sequencing (RNA-Seq) to profile the whole genome expression of mangrove in response to salt stress in order to identify tolerance-related genes (Figure 1). Additionally, functional genomics tools can be used to isolate and characterize mangrove genes involved in plant tolerance to the abiotic stresses (Figure 1). In both techniques (Figure 1), one-month-old A. marina seedlings will be grown hydroponically in half-strength MS basal medium amended by Nacl for salt stressor polyethylene glycol (PEG) for drought stress while control plants will grow in a nutrient solution. Root and leaf samples should be collected at early time points and before the appearance of severe symptoms of stress.
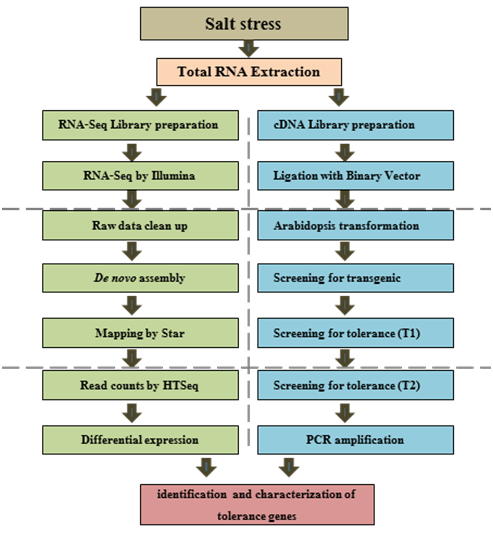
Figure 1 Exploring the genetic repertoire of mangrove using different research strategies for further improvement of agronomic crops. The first strategy uses RNA-Seq approach to identify tolerance-related genes while second strategy focuses on using a high-throughput functional screening by overexpression the whole cDNA library of mangrove in Arabidopsis thaliana as a sensitive species for the salt stress.
Extracted RNA would be used to construct cDNA libraries that would further be used either for RNA-Seq or overexpression in Arabidopsis thaliana:
Both techniques (Figure 1) have advantages and disadvantages. The final decision about the suitable technique depends on the finical situation and experience of each lab. With the advanced technology of high throughput sequencing, the cost for RNA-Seq is decreasing; however, this cost is higher than the cost of functional genomics. RNA-Seq can cover the whole genome of particular species enables scientists to identify regulatory genes such as transcription factors. One significant challenge facing RNA-Seq is the availability of reference genome and the computational ability to handle a big data. On the other hand, the cost and the demand for bioinformatics decrease when functional genomics is used. Additionally, there is no further need to use other functional genomics to confirm the obtained results. A major disadvantage of functional mining is the potential of missing important genes. This is more likely either during cDNA preparation or while screening transgenic plants. Additionally, the time and space required for conducting the functional mining are considered disadvantages. The following table summarizes advantages and disadvantages of each technique (Table 1).
Comparison |
RNA-Seq |
Functional Miming |
Cost |
High |
Less |
Bioinformatics use |
High |
Less |
Whole Genome Covering |
Yes |
No |
qRT-PCR for Data Validation |
Yes |
No |
Time |
~6 months |
~12-18 months |
Missing Important Genes |
No |
Yes |
Future Confirmation using Functional Genomics |
Yes |
No |
Table 1 Summarizes a comparison between RNA-Seq and functional miming of mangrove cDNA in other plant species
Some plant species are able to grow in arid areas making them important subsistence crops in extremely arid areas. Date palm (Phoenix dactylifers L.), is a tree that can be cultivated in hot and dry regions of the world, yet still produces high yields. This tree plays significant roles related to economy, society and environment. Additionally, the date fruits are rich in proteins, vitamins and mineral salts, providing the human body with essential nutrients. However, date palm tree is not among halophyte species, it is able to survive under drought and salinity stresses. This ecological survival makes its genome a unique repertoire for genes that may be involved in abiotic stress tolerance. While tolerance mechanisms to abiotic stress in the model and agronomic crops have been investigated using different genomic tools, no such study has been reported in date palm. It is important to conduct future research projects to explore the mechanisms of date palm tolerance to salinity and drought stresses and to identify genes that might further be used for the genetic improvement of plant species that are sensitive to the salinity and drought stresses.
Our results (Osman Radwan, unpublished data) from the genome expression of date palm using RNA-Seq in response to salinity and drought stresses revealed a share signaling pathways between drought and salt stress. One example is the activation of ABA signaling pathways through SNF1-related protein kinases 2 (SNRK2), which phosphorylates ABA-responsive element binding factors, inducing their transcriptional activity. Another example of triggering tolerance mechanisms in date palm is the activation of transcripts involved in synthesizing and transporting proline such as proline-rich protein, proline transporter, amino acid transporter and amino acid permease. Additionally, certain key genes involved in sodium uptake and transport are down-regulated suggesting a potential mechanism under salinity stress to slow down up-take and transportation salt solutes inside plant tissues (Osman Radwan, unpublished data). These promising results inspire us for future research studies in date palm using available genomics and functional genomic tools. Several recent studies have demonstrated that the adaptation of some plants to severe environmental conditions is attributed to genetic abilities of their associated microbes.16 For example, all plants in natural ecosystems are thought to be symbiotic with mycorrhizal and/or endophytic fungi reflecting fitness benefits conferred by fungi. These symbiotic relationships contribute to or are responsible for plant adaptation to the stress.17,18 Additionally, several studies focused on the identification of bacteria that enhance plant tolerance to stress include plant growth-promoting bacteria (PGPB), symbiotic bacteria and bacteria containing 1-aminocyclopropane-1-carboxylate deaminase.19‒21 Therefore, it is important to identify plant-associated microbes that may further be used to improve plant tolerance to abiotic stress and to understand mechanisms underlying enhanced plant tolerance.
Metagenomics is a new powerful technology tool with potential applications to solve practical challenges in medicine, engineering, agriculture, ecology and sustainability. It is widely used to better understand potential changes in microbial community structure in response to environmental changes. Metagenomics can also be used to address the role of the plant-associated microbiome in supporting plant tolerance to biotic and abiotic stresses. In addition to extensive genomic analysis, this approach can target specific sequence regions such as 16S rRNA (microbial prokaryotic) or 18S rRNA (microbial eukaryotic). It is an effective way to identify plant-associated microbiomes that may play an important role in supporting plant tolerance to biotic and abiotic stress. Microbial communities associated with plants provide a wide variety of ecosystem services necessary for plant growth such as fixation of atmospheric nitrogen, nutrient cycling, disease suppression and sequestration of essential metals. Functional metagenomics strategies can be used to explore the interactions between plants and microbes through a culture-independent study of these microbial communities.22 Additionally, a metagenomics approach can improve cropping systems by coupling the relationship between plants and their associated microbes, which will lead to enhancing crop health.
Cellular dedifferentiation also contributes to plant responses to abiotic and biotic stresses. During stress response, a select group of transcription factors alter gene expression; lower the overall amount of produced protein and rearrange chromatin structure. These processes are trademarks of cells that are dedifferentiated and upon introduction of stress; cells enter a stem cell-like state while awaiting new programming.23 Small RNAs may regulate plant stress dedifferentiation as they are known to do in animal systems. It has already been shown that small RNAs are involved in the plant response to abiotic stress.24‒26 Argonaute 1, Dicer-like 1 and micro RNAs targeting these two genes have altered expression levels in plants undergoing water stress.27,28 These two genes were found to be essential for cells maintaining a stem cell-like state.29,30 In high-throughput sequencing study, a total of 71 and 50 Glycine max micro RNAs were differentially expressed under osmotic and saline stresses, respectively.31 Further understanding the role of cell dedifferentiation and small RNAs in stress response may provide new targets for creating multi-stress tolerant cultivars.32‒37
Locating areas of abiotic stress as well as collecting and mapping spatial distributions of crops will help manage stressed areas for improvements. Geospatial tools such as remote sensing; geographical information system (GIS) and global positioning system (GPS) have the capabilities to fill gaps in plant stress analysis. Increased use of GIS and GPS and other emerging spatial data acquiring technologies such as high resolution airborne or space borne sensors can now provide accurate data and processing techniques to extract a variety of crop condition related information. Types of applications include crop condition assessment, yield estimation, crop type classification, soil characteristic mapping, soil type, soil erosion, moisture changes and soil management practices. The information can then be used to develop and implement spot plans that optimize the use of fertilizer and water. Other strategies to improve the tolerance of agronomic crops to abiotic stress include functional, structural and comparative genomics approaches. Transcriptomic studies in different plant species revealed important roles of some signaling pathways: abscisic acid (ABA) responders, expansins, heat shock proteins, inositol phosphates, lipases, metallothioneins, ROS scavengers, isoflavonoids, polyamines and transcription factors (bZIP, MYB, NAC and WRKY families).38‒43 Calcium signaling is involved in various stress responses and calmodulin proteins act as sensors to changes in Ca2+ concentration.44 Overexpression of the soybean SCaM-4 in A. thaliana was found to confer high-salt tolerance via the activation of an MYB2 transcription factor.45 Leucine-rich repeat receptor-like protein kinases (LRR-RLKs) have been indicated in both disease resistance46 and abiotic stress response.47 In soybean, GM-RLK3 is a receptor-like kinase gene, which shares similarity with A. thaliana BAM1 gene suggests a similar functional role in promoting meristem size and cell differentiation. Individual transposon-insertion mutant alleles of BAM1 have shown resistance to NaCl and mannitol stress.48 Stable transgenic plants were developed using immature cotyledon explants to study the functional role of this gene in soybean (Osman Radwan, unpublished data). Data analysis from RNA-Seq showed that overexpression of Gm-RLK3 triggers the transcription of other members of RLK and different signaling pathways involved in phosphorus, nitrogen and carbon assimilation (Osman Radwan, unpublished data). It is necessary to characterize genes that, when manipulated in the root system, will contribute to the tolerance of abiotic stress. By engineering plants with specific alterations in gene expression, we hope to mitigate yield losses experienced by plants in these environmental extremes.
As a first step of functional genomics, a list of genes needs to be selected from published expression studies focused on plant responses to abiotic stress. Datasets from different resources can be combined for meta-analysis to identify genes with expression changes that are consistent over multiple studies for each stress category. To further round out the list, transcript-level changes would be compared with published proteomics studies raising the priority of a given candidate. The second step of functional genomics should focus on verification of gene expression in tolerant and intolerant lines under stress, which will provide a list of genes that specifically respond to abiotic stress. Once the gene expression profiles are confirmed, the functional genomic tool would be used to assess the functional role of each gene through developing constructs of selected candidate genes driven by a root‐specific promoter and/or ubiquitous promoter. This is important because overexpression of the target gene in the root was able to increase yields under drought, but overexpression in the whole plant did not produce this phenotype.49 Adverse pleiotropic effects of gene overexpression in roots could cover any positive effects the gene has in the roots. Therefore, root-specific promoters linked to GFP should be tested to select root-specific promoters. Some of root-specific promoters are: Cryptic T80,50,51 Pyk10,52 RCc349 and SIREO.53 Because the stable transformation of some agronomic crops like soybean is laborious, a hairy root transformation could be used as an alternative method.54 Hairy root transformation may offer clues as to how each gene is affecting root architecture or root growth behavior to mediate that tolerance. Constructs used to develop hairy roots that show improved tolerance to abiotic stress will be used to generate stable transgenic lines using Agrobacterium tumefaciens.
Genetic engineering the plant signaling pathways would be a valuable option to improve tolerance of agronomic crops to abiotic stress. For example, ethylene produced by host legumes negatively regulates nodulation of rhizobia. On the other hand, rhizobia overcome this negative effect, either by producing 1-aminocyclopropane-1-carboxylate (ACC) deaminase which degrades ACC (a precursor of ethylene), or by producing rhizobitoxine, which inhibits ethylene synthesis.55 Because ethylene is induced during abiotic stress, one possibility to increase plant tolerance to abiotic stress is the genetic engineering of ethylene pathway. This can be done by using genetic engineering tools to either block or inhibit ACC, a key enzyme in the ethylene biosynthesis pathway. One strategy is to over express genes originating from rhizobia species controlling the production of ACC deaminase and rhizobitoxine. This approach should lead to degradation or inhibition of ACC thereby increasing tolerance to abiotic stress and enhanced nodule production in legume plants. In addition to using the available genomics and functional genomics tools in agronomic crops, the comparative genomic approach can be implicated to translate the discoveries made in the plant model systems. One approach to achieving this research target would be to explore the homologous genes from model systems such as A. thaliana and Medicagotruncatula and use the vast collection of mutants. Genomic information developed from model plants would be useful for identifying genes that control similar biological and physiological processes in agronomic crops.
Harsh conditions of drought and salinity affect the growth of plants leading to limited agricultural land and yield loss, which eventually demands sustainable solutions. Necessary steps should be taken to overcome these problems to increase the efficiency of agronomic crops within cultivated areas. Developing tolerant plants through better understanding tolerance mechanisms and exploring new genetic repertoires from natural and desert ecosystems would be a priority to improve tolerance in agronomic crops. Metagenomics can be used to explore the diversity and functional roles of plant microbiomes that support plant tolerance to abiotic stress. Additionally, the process of identifying, quantifying and mapping abiotic stress is compulsory for efficient allocation of environmental stresses and development of improved crop management strategies. Altogether, multidisciplinary approaches of plant ecology, microbiology, genomic and geospatial tools would be integratedin developing new strategies to improve plant tolerance to abiotic stress to increase the sustainable food supply.
This research was made possible by a grant from the Qatar National Research Fund (QNRF) under National Priorities Research Program (NPRP-5-1040-4-013). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the QNRF. I would like to thank Caroline Keller for the critical reading of the manuscript.
Author declares that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.