Journal of
eISSN: 2373-6453


Antiretroviral Therapy is classified in six classes and acquired great achievements in HIV inhibition, but this advantage can be affected by developing drug resistance. The combinations of antiretroviral (ARV) drugs have been proven as effectiveness key to control the progression of AIDS; however, these benefits may be compromised by drug resistancy. Thus, drug-resistance testing has become an important tool to the management of HIV-infected patients. Viral RNA was extracted from the plasma by using the QIAamp→ Viral RNA kit (Qiagen, Germany) according to the manufacturer’s protocol. RT-PCR performed with the one-step PCR kit (Qiagen, Germany) and Nested PCR.
We used the Taq Polymerase master mix kit (Sinaclone, Iran) to amplify of target gene, then products sequenced and analyzed in Stanford HIV drug resistant database. 25 patients, including 13 (52%) males and 12 (48%) females that all of whom received antiretroviral treatment and data indicated that 19 of 25 patients (76%) had RT mutation that consequently reduce susceptibility to NRTI and NNRTI and 4 patients (16%) had PR mutations. In this study two patients have mutation in L100 combine with K103N and 3% of patients have mutation in T215 that mutation in T215, suggesting that they had likely been infected originally with a mutant virus. The mutation rate in RT gene was 76% which may be due to not adherence to the treatment regimen by the patients may cause drug resistancy.
Keywords:Antiretroviral, Human immunodeficiency virus, Drug resistancy, ART, Mutations, HIV
ART, Antiretroviral Therapy; NNRTI, Non-Nucleoside Reverse Transcriptase Inhibitor; NRTI, Nucleoside/N Ucleotide Reverse Transcriptase Inhibitor; PI, Protease Inhibitor; AZT, Zidovudine; D4T, Stavudine; DDI, Didanosine; TDF, Tenofovir Disoproxil Fumarate; EFV, Efavirenz; NVP, Nevirapine; ETR, Etravirine; LPV/R, Lopinavir and Ritonavir; SQV, Saquinavir; IDV, Indinavir; NFV, Nelfinavir; ATV, Atazanavir; DRV, Darunavir; FAPV, Fosamprenavir; TPV, Tipranavir
Antiretroviral Therapy is classified in six classes and acquired great achievements in HIV inhibition, but this advantage can be affected by developing drug resistance.1,2 The use of HAART is instrumental in controlling the development of drug resistance where the drug resistance was more frequent in the individuals who started treatment in the 1990s with single ART regimen.3 The combinations of antiretroviral (ARV) drugs have been proven as an effectiveness key to control the progression of AIDS; however, these benefits may be compromised by drug resistancy. Thus, drug-resistance testing has become an important tool to the management of HIV-infected patients. In Iran, the first line of antiretroviral regimen is zidovudine (AZT), lamivudine (3TC) and efavirenz (EFV).4 Counseling and medicines are free of charge for HIV positive people in Iran. Drug resistance creating as a result of virus genome mutation creation to escape the harmful effects of drug and inadequate suppression of viral replication. HIV virus can be resistance to all of the invented antiretroviral drugs. Drug resistance develops unavoidable and can transmit to other populations.5 In each viral life cycle, plenty of mutations happen that ensure the patient has a divers mixture of viruses that are different at the type and amount of mutations.6
The prevalence rate of drug resistance mutations depends on three factors, the selective advantage that confer by mutations, the prevalence of the mutant in viral population and the level of ART drugs.7 Drug resistance can occur in a patient who receiving ART regimen or transmitted to a naïve one, from the other individual who carried the drug resistance mutations.8 The studies show drug resistances have been emerged in 10% of individuals who used ART after 2 years and treatment of 30% of individuals with at least one drug resistance mutation have been failed after 6 months.9
Treatment failure happens when the medication can’t control the infection and categorizes in 3 groups. Virologic failure which happens when viral load doesn’t decrease while taking medication, Immunologic failure which happens when immune system doesn’t response to medication and CD4 count doesn’t increase and clinical progression which happens when the person despite taking medication has clinical signs.10 According to emerge of drug resistance testing in HIV-infected patients who receive ART, in treatment planning and directing the medical policy of the countries the aim of this study is to evaluate protease and reverse transcriptase region drug resistance in HIV-infected people with treatment failure by genotyping method.
Ethics statement
This study was approved by the institutional ethics review board of volunteers from whom specimens were obtained provided written informed consent.
Sample collection
Sample collection: 25 HIV positive individuals with treatment failure (individuals that takes therapy, with clinical signs and declining CD4) who were referred to the behavioral disease department of Imam Khomeini hospital (Tehran, Iran) with the mean CD4 215 were selected and informed consent was taken from them. 10(40%) of specimens were taken Kaletra base drugs, 6(24%) of specimens were taken NVP base drugs and 9(36%) of specimens were taken EFV base drugs. The blood samples were collected in sterile EDTA containing tubes and plasma was collected and stored at -80°C.
RNA Extraction and RT-Nested PCR
Viral RNA was extracted from the plasma with use the QIAamp→ Viral RNA kit (Qiagen, Germany) according to the manufacturer’s protocol. RT-PCR performed with the one-step PCR kit (Qiagen, Germany). 1 µl (10 Micro Mole/μl) of sense and antisense primers (Table 1) was made for cDNA synthesis. For the RT region, viral RNA was pre-denatured at 50°C for 30 min and 95°C for 15 min, 38 cycle of denaturation at 94°C for 30 sec, annealing at 53°C for 35 sec, extension at 72°C for 1 min and final extension temperature of 72°C for 5 min and for Pr region, pre-denaturation at 50°C for 30 min and 95°C for 15 min ,38 cycle of denaturation at 94°C for 30 sec, annealing at 55°C for 35 sec, extension at 72°C for 1:10 min and final extension temperature of 72°C for 5 min.
Protease |
Sequence primer |
Position |
Outer primers |
||
Prot. F 1 |
5’-TAATTTTTTAGGGAAGATCTGGCCTTCC-3’ |
2082 to 2109 |
Prot. R2 |
5’-GCAAATACTGGAGTATTGTATGGATTTTCAGG-3’ |
2703 to 2734 |
Inner primers |
||
Prot. F 3 |
5’-TCAGAGCAGACCAGAGCCAACAGCCCCA-3’ |
2136 to 2163 |
Prot. R 4: |
5’-AATGCTTTTATTTTTTCTTCTGTCAATGGC-3’ |
2621 to 2650 |
Reverse Transcriptase |
||
Outer primers |
||
RT1.F |
5’-TTTYAGRGARCTYAATAARAGAACTCA-3’ |
2790-2818 |
RT2.R |
5’-CCTCITTYTTGCATAYTTYCCTGTT -3’ |
3615-3590 |
Inner primers |
||
RT3.F |
5-TTYTGGGARGTYCARYTAGGRATACC-3’ |
2823- 2850 |
RT4.R |
5’-GGYTCTTGRTAAATTTGRTATGTCCA-3 |
3601-3575 |
Table 1 Sequence of Primers
For Nested PCR, we used the Taq Polymerase master mix kit (Sinaclone, Iran). Master Mix prepared 0.5 µl (10 MicroMolar/μl) with sense and antisense primers11,12 the cycling conditions for RT region were as follows: one round of 94°C for 3 min, 38 cycles of 93°C for 30 sec, 53°C for 35 sec and 72°C for 1 min, with a final extension of 5 min at 72°C. For the Pr region, we used the same cycling conditions but in annealing we used 55°C instead of 53°C for 35 sec. The PCR product sequenced via Sanger Sequencing method by ABI 3730 XL (96 capillaries) system.
Drug resistance analysis
The sequences were edited by BioEdit and Chromas software and were analyzed with the Stanford University HIV Drug Resistance Database.
The 25 Individuals of this study included 13 (52%) males and 12 (48%) females that all of whom received antiretroviral treatment. Of the 25 amplified sequences, resistance to the protease inhibitors was observed in 4 (16%) of the sequences and to reverse transcriptase inhibitors in 19 (76%) sequences. 5 (20%) of the specimens were infected by viruses with low resistance to NRTI drugs, 5 (20%) of low resistance to NNRTI and 5 (20%) with low resistance to PI drugs. NRTI intermediate resistance identified in 6 (24%) and NNRTI and PI intermediate resistance identified respectively in 4 (16%) and 6 (24%) of specimens.
The highest level drug resistance was observed in ETR in 17(68%) of patients and after that in 3TC and FTC with 16(64%) frequency (Table 2). M184V and T215F were the most frequent NRTI resistance mutations and observed in 15(60%) and 9(36%) of patients respectively. G190A 10(36%) and L10I 2(8%), respectively were the most frequent NNRTI and PI resistance mutations. Clinically, K103N was the most important drug resistance mutation to NNRTI and was observed in 2(8%) patients (25). Table 3 describes the drug resistance mutations in the PR and RT region.
N RTI |
High (%) |
Intermediate (%) |
Low (%) |
3TC |
16 (64%) |
0 |
0 |
ABC |
3 (12%) |
10 (40%) |
3 (12%) |
AZT |
6 (24%) |
3 (12%) |
1 (4%) |
D4T |
6 (24%) |
4 (16%) |
2 (8%) |
DDI |
4 (16%) |
6 (24%) |
4 (16%) |
TDF |
0 |
2 (8%) |
10 (40%) |
NN RTI |
High (%) |
Intermediate (%) |
Low (%) |
EFV |
9 (%) |
8 (32%) |
2 (8%) |
ETR |
17 (%) |
8 (32%) |
10 (40%) |
NVP |
5 (%) |
1 (4%) |
1 (4%) |
RPV |
0 |
5 (20%) |
8 (32%) |
PIs |
High (%) |
Intermediate (%) |
Low (%) |
ATV/r |
1 (4%) |
2 (8%) |
2 (8%) |
DRV/r |
0 |
0 |
0 |
FPV/r |
1(4%) |
1(4%) |
0 |
IDV/r |
1(4%) |
1(4%) |
0 |
LPV/r |
1(4%) |
1(4%) |
0 |
NFV |
1 (4%) |
1 (4%) |
1 (4%) |
SQV/r |
0 |
1 (4%) |
2 (8%) |
TPV/r |
1(4%) |
0 |
1 (4%) |
Table 2 Number of high, intermediate and low drug resistance to reverse transcriptase and protease inhibitors drugs classes among Iranian HIV-infected patients
ART, Antiretroviral Therapy; NNRTI, Non-Nucleoside Reverse Transcriptase Inhibitor; NRTI, Nucleoside/N Ucleotide Reverse Transcriptase Inhibitor; PI, Protease Inhibitor; 3TC, Lamivudine; FTC, Emtricitabine; AZT, Zidovudine; D4T, Stavudine; DDI, Didanosine; ABC, Abacavir; TDF, Tenofovir Disoproxil Fumarate; EFV, Efavirenz; NVP, Nevirapine; ETR, Etravirine; LPV/R, Lopinavir And Ritonavir; SQV, Saquinavir; IDV, Indinavir; NFV, Nelfinavir; ATV, Atazanavir; DRV, Darunavir; FAPV, Fosamprenavir; TPV, Tipranavir
NRTI mutations |
N (%) |
NNRTI mutations |
N (%) |
PI mutations |
N (%) |
M41L |
1(%4) |
V90I |
1(%4) |
L10F/I |
3(%12) |
D67N |
1(%4) |
L100I |
1(%4) |
V32I |
1(%4) |
T69N |
1(%4) |
K101E |
2(%8) |
L33F/I |
2(%8) |
K70R |
3(%12) |
K103T/N/Q |
4(%16) |
M46I |
1(%4) |
L74V |
1(%4) |
V106I |
1(%4) |
I47M/A |
2(%8) |
V75M/L |
2(%8) |
V108I |
1(%4) |
I54V |
1(%4) |
Y115F |
1(%4) |
E138G |
1(%4) |
Q58E |
1(%4) |
M184V |
15(%60) |
V179T |
2(%8) |
A71V |
1(%4) |
L210W |
3(%12) |
Y181C |
2(%8) |
T74S |
1(%4) |
T215Y/S/F |
11(%44) |
Y188F/L |
3(%12) |
V82A |
1(%4) |
K219W/E/Q/R |
6(24) |
G190A/T |
11(%44) |
L89V |
1(%4) |
H221Y |
1(%4) |
||||
F227L |
1(%4) |
||||
M230L |
2(%8) |
||||
K238T |
1(%4) |
||||
Y318F |
1(%4) |
||||
Any NRTI |
N (%) |
Any NNRT |
N (%) |
Any PI Mutations |
N (%) |
1 Mutation |
13(%52) |
1 Mutation |
13(%52) |
1 Mutation |
13(%52) |
2 Mutations |
1(%4) |
2 Mutations |
6(%24) |
2 Mutations |
1(%4) |
≥3 Mutations |
5(%20) |
≥3 Mutations |
1(%4) |
≥3 Mutations |
0 |
Table 3 Number and percentage of protease and reverse transcriptase drug resistance mutations had been observed in this study based on Stanford HIV Resistance Database
ART, Antiretroviral Therapy; NNRTI, Non- Nucleoside Reverse Transcriptase Inhibitor; NRTI, Nucleoside/ Nucleotide Reverse Transcriptase Inhibitor; PI, Protease Inhibitor
To better understand the molecular epidemiology of HIV infection, first we enter the reverse transcriptase sequence (RT) into the mega6 software that before analyzed in the HIV DRUG Resistant database by adding Iranian and full sequence CRF35-AD genome reported from a neighboring country, Afghanistan and reference sequence of M, N, O, P as out-groups that existed in the HIV database then we select Clustal W and aligned sequence. After alignment we drew the phylogenetic tree with mega6 and RDP4 software. We used neighbor going method with 1000 bootstrap. 17 of our isolated belong the cluster, Of AD 35 and one belong cluster A and Other made cluster AE and C. Almost all of 17 CRF35-AD candidates showed the highest similarity to the CRF35-AD reference genome that belongs to Iran and Afghanistan sequence that before reported and excited in NCBI and HIV database that showing in Figure 1.
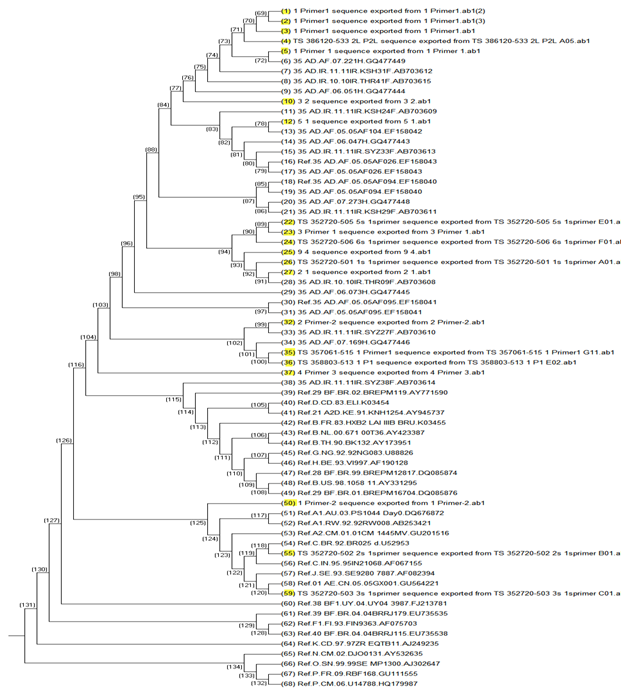
Figure 1 Phylogenetic tree analysis of Iranian HIV-1 isolates.
The tree was constructed with HIV-1 RT genome sequences. Our sequences are represented with highlighted yellow color, and subtype reference isolates are represented by their subtype and name. HIV-1 group O, M, N isolate, was used as the out-groups.
We investigated 25 HIV-infected patients with treatment failure included of 13 (52%) males and 12 (48%) females. In this study, we described the molecular characteristics of a part of HIV-1 which involves RT and Pr genomic region. HIV genome was isolated from patients who were receiving HARRT under the supervision of behavioral medicine clinic (Immam khomeini hospital Tehran-Iran). In agreement with previous studies in Iran, most of the patients receiving antiretroviral therapy with the detectable virus had evidence of drug resistance mutation.13 Many of patients were resistance to multiple drug classes and many of these mutations elaborate resistancy to other drugs in the same class.
This data indicated that 19 of 25 patients (76%) had RT mutation that consequently reduces susceptibility to NRTI and NNRTI and 4 patients (16%) had Pr mutations. The choice of the treatment regimen for these patients was the combination of reverse transcriptase inhibitor, zidovudine (AZT), lamivudine (3TC), efavirenze (EFV), and protease inhibitor Kaletra. There was 36% G190A mutation in that is a no polymorphic mutation selected by NVP and EFV.14,15 Occurrence mutation of G190A reduces NVP susceptibility more than 50-fold and EFV susceptibility 5 to 10-fold.16,17 It does not appear to be selected by ETR and RPV to reduce their antiviral activity.18 M184V detected in RT gene of 15 (60%) patients.
The M184VI is the most common NRTI-resistance mutations. Although they cause high-level in vitro resistance to 3TC/FTC, they are not contraindications to 3TC/FTC because they increase TDF, AZT, and d4T susceptibility and decrease viral replication fitness.19 We have one patient who has mutations in T215F/S/Y. The previous study indicated T215S often can be detected in isolates that eventually go on to develop the drug-resistance mutations T215Y and T215F. Conversely, isolates that once had T215Y or T215F and are no longer exposed to nucleoside RT inhibitors often revert to T215C or T215D or T215E rather than to wild type.20
T215S, T215C, T215D, and T215E do not cause phenotypic nucleoside RT inhibitor resistance. However, these mutations indicate the presence of selective drug pressure and strongly suggest that a population of truly resistant viruses. The study that done on more than 600 infected patients, 3% viruses containing one of these T215 mutations, suggesting that they had likely been infected originally with a mutant virus.20 Because of that, this sample maybe originally infected with mutant virus (Transmitted Drug Resistance) or the resource viruses mutinied before of start drug treatment in this patient. 17 (68%) of 25 patients showed drug resistant for ETR.
The mutation in L100I is selected in patients receiving EFV, ETR, and RPV.15 Although etravirine use was not reported in Iran, accumulation of NNRTI mutations among participants on EFV resulted in frequent high-level (68%) or low-level (40 %) cross-resistance to ETR.21,22 The phylogenetic tree represented highest similarly between our RT sequence and CRF-AD35 and this study correlated with other study was done in Iran23 exempt subtype C that this sample belonged east of Iran(Mashhad). Subtype C is prevalence in India24 and historical of patient represented that he had traveled to India. In the final one of the advantages of drug resistance assay in HIV-infected patients is that this assay consequently causes to detect the mutations that occurred in circulated viruses and as a consequence caused treatment failure in patients. Drug resistance assays have been done and base on the results next treatment regimen have been chosen.
In present study mutation rate in RT gene is about 76%. The reason may be the lack of adherence to treatment. The monitoring of patients with treatment failure who are under treatment has substantial benefits in terms of survival and quality of life for these patients.
This study was supported by Iranian Research Center for HIV AIDS (IRCHA) Iranian Institute for Reduction of High-Risk Behaviors Tehran University of Medical Sciences (TUMS). That provided sample and grants of this study.
None.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
 World Malaria Day, observed annually on April 25SUP>th. The purpose of this day is to raise awareness about the global struggle to combat malaria, highlighting the need for continued research, prevention, and treatment strategies to eradicate this deadly disease. So, it is an opportunity to all the researchers to submit your interesting papers on raising the approachability on modern vaccine development, advances in disease control, prevention and eradication and the submissions received till April 25th, 2024 will be offered a best discount of 30% for publication in Journal of Human Virology & Retro virology.
World Malaria Day, observed annually on April 25SUP>th. The purpose of this day is to raise awareness about the global struggle to combat malaria, highlighting the need for continued research, prevention, and treatment strategies to eradicate this deadly disease. So, it is an opportunity to all the researchers to submit your interesting papers on raising the approachability on modern vaccine development, advances in disease control, prevention and eradication and the submissions received till April 25th, 2024 will be offered a best discount of 30% for publication in Journal of Human Virology & Retro virology.