Journal of
eISSN: 2373-6453


HIV-1 Tat-mediated transactivation of the long terminal repeat (LTR) requires an interaction with the transactivation response element (TAR) and is facilitated by NF-κB and Sp factors binding to sequences upstream of the transcriptional start site. Studies conducted pre- and post antiretroviral therapy identified HIV-1-infected patients harboring a C-to-T change at position 5 of the consensus B sequence of Sp binding site III (a knockout configuration with respect to binding of Sp1). HIV-1 LTRs and Tat were amplified and cloned from patients in the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort. As one simple approach to examine the impact of sequence variation on LTR fitness, LTR clones from a single patient (patient 19) were used in transient expression studies in both T-cell and monocyte-macrophage cell lines. These results demonstrated that one of 4 clones could not be transactivated by Tat derived from laboratory strain IIIB or from the patient. After inspection of the sequence of the patient-derived LTR clone defective with respect to Tat-mediated transactivation, additional alterations within the LTR were identified in a number of transcription factor binding sites in the LTR core region as well as in the TAR element that suggested that these sites may also contribute to the Tat non responsiveness. In this regard, site-directed mutagenesis indicated that optimal Tat-mediated transactivation depended on both position 32 of the TAR element and binding of Sp to all three Sp binding sites within the LTR. These studies indicate that a vast majority of LTRs derived from the integrated provirus in the peripheral blood compartment of a number of infected patients maintained the ability to drive basal and Tat-mediated transcription but that just a small number of nucleotides were shown to have a detrimental impact on LTR activation in both T cells and cells of the monocyte-macrophage lineage.
Keywords:HIV-1, LTR, TAR, Tat, Transactivation, Genetic variation, Sp binding sites
The production of human immunodeficiency virus type 1 (HIV-1) RNA and protein subsequent to infection is achieved using the integrated provirus as a template to guide production of both regulatory and structural gene products. The molecular diversity of HIV-1 is a critical mediator of viral replication and overall viral fitness.1 HIV-1 sequence variation within the LTR and other viral genes, such as env, Tat, and others, impacts viral gene expression and replication as well as viral tropism, which may affect overall HIV disease severity based on the relative fitness of the virus. Located at both ends of the integrated genome are two identical viral long terminal repeat (LTR) sequences, with the 5′ LTR serving as the promoter for HIV-1 transcription and the 3′ LTR providing for nascent viral RNA polyadenylation. The HIV-1 LTR is approximately 640 bp in length and is divided into the unique 3 (U3), repeat (R), and unique 5 (U5) regions. The U3 region is further subdivided into the core, enhancer, and modulatory regions and their associated transcription factor binding sites that guide the DNA–protein and protein–protein interactions important for LTR activity and regulated viral gene expression. The core region is required for HIV-1 basal transcription and is positioned approximately from 20 to 60 bp upstream of the transcriptional start site. The core region includes the TATAA element and three GC rich binding sites for the Sp family of transcription factors. The TATAA box binds the TATAA binding protein (TBP) and other host proteins that comprise the RNA polymerase transcription complex.2 The enhancer region is located upstream of the GC box array and consists of two tandem nuclear factor-kB (NF-kB) binding sites. The modulatory region, located further upstream of the NF-κB sites, includes binding sites for CCAAT/enhancer binding proteins (C/EBP), activating transcription factor/cyclic AMP response element binding (ATF/CREB) factors, lymphocyte enhancer factor (LEF-1), nuclear factor of activated T cells (NFAT), and a number of other transcription factors.3,4
HIV-1 Tat-mediated transactivation of the LTR requires the interaction of Tat with a unique complementary RNA regulatory segment of the LTR designated the transactivation response element (TAR), which is located at nucleotides +1 to +59 within the R region of the HIV-1 LTR.5 TAR forms a stable stem-bulge-loop structure like a hairpin, including a six-nucleotide loop, a trinucleotide pyrimidine bulge, and an extensive duplex structure.6 The bulge structure (U23C24U25 in most quasi species) is critical for the interaction of TAR with the Tat protein.7 The Tat protein alone binds only to the TAR bulge, and not to the loop structure. However, the loop structure (+30 to +35) can affect Tat transactivation activity because the loop affects the interaction of Tat with other proteins, such as cyclin T1 that in turn can influence the transactivational activity of Tat.8 Cyclin T1 within the P-TEFb complex (positive transcription elongation factor) interacts with the activation domain of Tat and binds to the central loop of TAR.9 Once cyclin T1 binds to Tat, the CycT1–Tat complex is able to bind both the bulge and loop region of TAR with a higher affinity than Tat alone, thereby forming the CycT1-Tat-TAR ternary complex.9-11 In the bulge region of TAR, residue U23 has been shown to be essential for the Tat–TAR interaction, whereas the other two residues C24 and U25 can be replaced by other nucleotides without affecting Tat binding.12-14 The two bp above the bulge (G26-C39 and A27-U38) and one below (A22-U40) have also been shown to contribute to Tat binding.12-14
The HIV-1 genotype and resultant phenotype are important variables with respect to viral gene expression, replication, and fitness of specific viral quasi species during the course of HIV disease in the immune and central nervous systems.15 Sequence alterations within the Sp elements in the core region of the LTR can affect basal, activated, and Tat-mediated LTR activation. In this regard, deletion of the LTR Sp binding sites or GC box array has been shown to dramatically reduce Tat transactivation.16 In clinical samples derived from HIV-1-infected patients in years prior to or after the implementation of effective combination antiretroviral therapy(cART), specific single-nucleotide polymorphisms have been observed within the NF-κB-proximal Sp transcription factor binding sites that exhibit a very low affinity for their cognate factor and alter LTR-driven viral gene activation.17,18 Specifically, these studies reported that LTR sequence variants within the Sp site III (5T; a C-to-T change at position 5 of the binding site) were prevalent during the course of HIV-1 infection and exhibit very low binding affinities for members of the Sp transcription factor family.19,20
Understanding if proviral DNA in PBMCs of patients is defective and/or has the ability to be induced and replicate has become a major issue in understanding if viral eradication is a possibility. Recent studies have shown that there exist replication competent non-induced proviruses in the latent viral reservoir.21 Other studies have suggested that these proviruses are defective in replication and that this may be due to APOBEC3G editing in HIV proteins22 or due to integration sites.23 Given these observations and a growing interest in HIV-1 LTR genetic variation and its impact on HIV-1 transcription, we have examined naturally occurring sequence variation in the LTR to determine what variants may lead to a defective LTR in proviruses present within the peripheral blood and so determined the nature of the defective LTRs. In recent studies with HIV-1-infected patients in the cARTera, the previously identified low Sp affinity 5T variation has been shown to remain prevalent (24.9%), and continued investigations have explored the relative fitness of patient-derived peripheral blood HIV-1 LTRs containing the 5T Sp site III sequence configurations.24,25 These studies demonstrated that transient and stable expression assays of patient–derived LTRs containing the 5T Sp site III configuration as well as many other configurations of this binding site identified within the PBMC compartment performed in a number of T-cell and monocyte cell lines exhibited a wide range of functional activity, but that they were functional. In fact, studies have shown that the HIV-1 LTR may evolve in a compartmentalized manner, suggesting that genetic variation within the HIV-1 LTR plays a role in adaptation of the virus to different tissues such as the CNS.26,27
During the course of these analyses, patient 19 was identified as a patient who had several relatively functional LTR clones with but one exception. In this regard, a single HIV-1 LTR clone was identified that exhibited reduced basal transcriptional activity in T cells, and even more pronounced activity in monocytic cells, but was defective with respect to responses to Tat transactivation in both cell lines based on specific alterations in the TAR element and Sp binding sites. When the identified nucleotide variant located at nucleotide +32 of the TAR element was changed to the LAI nucleotide, Tat-mediated LTR transactivation was partially restored. In contrast, conversion of a number of other nucleotide variations observed within TAR of this particular LTR clone to LAI did not result in even partial restoration in the response to Tat transactivation. It was also demonstrated that restoration of the three Sp binding sites to the LAI sequence was necessary for further restoration of Tat-mediated transactivation. These studies indicate that just a small number of nucleotide sequence alterations can have a highly detrimental impact on Tat-driven LTR activation in both T cells and cells of the monocyte-macrophage lineage. This study also validates previous observations that have demonstrated cooperation between Sp binding and Tat/TAR interactions for LTR transactivation; although the studies reported here have suggested this cooperativity based on gain of function studies from a naturally occurring LTR derived from the PBMC compartment that was nonresponsive to Tat.
Patient 19-derived LTR from infected PBMC
Patients were recruited into the Drexel Medicine CARES Cohort as previously described.28,29 Forty milliliters of peripheral blood were drawn from each patient, of which 10 mL were used for drug testing. PBMCs were isolated from the remaining 30 mL using ficoll gradient (Ficoll-Paque Plus, Amersham Biosciences, and Piscataway, NJ). From the isolated PBMCs, genomic DNA was extracted from 5 × 106 cells using the QiagenDNeasy Genomic DNA isolation procedure (Qiagen, Valencia, CA). Nested PCR was then utilized to amplify HIV-1 LTRs from the isolated genomic DNA. HIV-1 primer 3 (5′-TGGAAGGGCTAATTTACTC-3′) and primer 4 (5′-TGACTAAAAGGGTCTGAGG G-3′) were used for the first round of amplification, and HIV-1 primer 5 (5′-CACTCCCAACGAAGACAAGA-3′) and primer 6 (5′-GAGGGATCTCTAGTTACC AG-3′) (Integrated DNA Technologies, Coralville, IA) were used for the nested amplification round. PCR-amplified LTRs were purified from 1% agarose gels using the Promega PCR gel purification procedure. Purified DNA was quantified using a Beckman Coulter (Brea, CA) DU800 spectrophotometer and then sent for sequencing (Genewiz). HIV-1 LTR sequences were aligned and analyzed for genetic variation within NF-kB, C/EBP, and Sp transcription factor binding sites as well as the TAR region using 4Peaks (http://nucleobytes.com/index.php/4peaks) and DNASTAR Meg Align software.
Cell Line Maintenance
The human T-cell line Jurkat (American Type Culture Collection, ATCC, TIB-152) and the human monocytic cell line U-937 (ATCC, CRL-1593.2) were grown in RPMI-1640 media (Roswell Park Memorial Institute) (from Cellgro, Herndon, VA). Media for Jurkat cells was supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gem Cell, Sacramento, CA), sodium bicarbonate (0.05%, Cellgro), and antibiotics (penicillin, streptomycin, and kanamycin at 0.04 mg/mL each; Cellgro). Media for U-937 cells was supplemented with 10% heat-inactivated FBS (Gem Cell), antibiotics (penicillin and streptomycin, at 0.04 mg/mL each, Cellgro), glucose (4.5 g/mL, Cellgro), sodium pyruvate (1 mM, Cellgro), and HEPES (10 mM, Cellgro). The cells were maintained at 37°C in 5% CO2 at 90% relative humidity.
Plasmid Construction and SDM
The LTR-containing DNA fragment (approximately 640 bp) was derived from the LAI molecular clone of HIV-1. The HIV-1 LAI LTR was PCR amplified using the forward primer: 5′-GGGGTACCTGGAAGGGCTAATTCACTCC-3′; and the reverse primer: 5′-TCCCCCGGGTGCTAGAGATTTTCCACA-3′ (from IDT). The underlined nucleotides indicate the restriction endo nuclease cleavage sites KpnI and SmaI, respectively. The amplified product was digested with KpnI and Sma I (Promega) and ligated into a modified pGL3-Basic vector, which contained the firefly luciferase (Luc) gene (Promega), to create the parental HIV-1-LAI-LTR-Luc expression construct. The parental construct was used as a template for SDM using the Quick Change Mutagenesis procedure as described by the manufacturer (Stratagene, La Jolla, CA) to produce the mutant construct LAI-LTR-SpIII-5T. The following primers were used for SDM, and the nucleotide that was mutated is underlined. The forward primer is 5′-CTTTCCAGGGAGGTGTGGCCTAGGCAG-3′ and the reverse primer is 5′-CTGCCTAGGCCACACCTCCCTGGAAAG-3′ (from IDT).
Construction of patient 19–derived HIV-1 LTR-driven luciferase vectors was performed as follows. First, the patient 19–derived LTRs were amplified using a forward primer LL8: 5′-GGGGTACCTGGAAGGGCTAATTTACTTCC-3′; and a reverse primer LL7: 5′-TCCCCCGGGCTAATTTACTCC-3′. Subsequently, the pGL3-Basic vector and LTR PCR products were digested with KpnI and XmaI (Promega). Once digested, the LTRs were ligated into the pGL3 vector and transformed into DH5k competent cells. Colonies were selected and DNA plasmids were obtained utilizing a mini preparation procedure as described by the manufacturer (Promega), then sequenced to confirm the maintenance of the original LTR sequence during the sub cloning procedure (Genewiz). Once the sequence was confirmed, a large-scale plasmid DNA preparation was utilized to perform transient transfection experiments to examine the functional properties of the LTRs in selected cellular phenotypes and with a number of cellular stimulatory conditions. Sequences were analyzed using Lasergene software from DNASTAR. All luciferase vector constructs were checked for purity by agarose gel electrophoresis and spectrophometric ratios at 260/280 and 260/230.
The parental 19-18 LTR construct was used as a template for SDM using the Quick Change Mutagenesis procedure as described by the manufacturer (Stratagene) to construct plasmids containing the 19-18 LTR including the mutated SpI, SpII, and/or SpIII binding sites changed to the LAI configuration, and/or TAR sequence with the base at positions 2, 32, or 52 of the TAR element changed to the LAI configuration. The following forward primers and their reverse complement primers (from IDT) were used for SDM, and the nucleotides that were mutated are underlined. The native patient 19-18 LTR clone is listed prior to the slash mark (/) while the mutated sequence is listed after the slash mark:
19-18 SpI 1A2A/Sp1 1G2G (LAI): CCTAGGCAGGACTGGGGAGTGGCGAGCC
19-18 SpII 2A6A/SpII 2G6G (LAI): GGAGGTGTGGCCTGGGCGGGACTAAGGAG
19-18 SpI, II/SpI, II (LAI): GAGGTGTGGCCTGGGCGGGACTGGGGAGTG
19-18 SpIII 5T/SpIII 5C (LAI): CTTTCCAGGGAGGCGTGGCCTAGGCAG
19-18 SpI, II, III/SpI, II, III (LAI): CTTTCCAGGGAGGCGTGGCCTGGGCGGG
19-18 TAR 2A/2G (LAI): GCGCTTGTACAGGGTCTCTCTGGTT
19-18 TAR 52A/52G (LAI): TCTGGCTGACTAGGGAACCCACTGC
19-18 TAR 32A/32G (LAI): AGATCTGAGCCTGGGAGCTCTCTGG
The pGL3-LTR recombinant plasmids were transformed into DH5k competent cells. Colonies were selected and DNA plasmids were obtained utilizing a mini preparation procedure as described by the manufacturer (Promega). The recombinant plasmids were sequenced to confirm the mutation(s) and maintenance of the original LTR sequence after completion of the SDM procedure (Genewiz). Sequences were analyzed using Lasergene software from DNASTAR. Once the sequence was confirmed, large-scale plasmid DNA was prepared as previously described and utilized to perform transient transfection experiments.
Amplification of Full-Length Tat from HIV-1-Infected Patient 19 4.4 kb Proviral DNA Clone
A two-step PCR amplification procedure was performed to obtain Tat exon I and II DNA from patient 19. In the first step, the two exons derived from patient 19 clone 4 were amplified in individual reactions utilizing the 4.4-kb HIV-1 proviral genomic fragment contained within the recombinant TA clones as template (primers F1 and R1 for exon 1 and primers F2 and R2 for exon 2, with primer R1 and F2 having an overlap region of 17 nucleotides). In the second step of the protocol, the two PCR products were ligated together using a mixture of the PCR products from the first step of PCR amplification as a template and using primer F1 and R2 for amplification. Because of sequence variation within different patient-derived HIV-1 Tat clones (especially between residues 60 to 72, which exhibited the greatest degree of genetic variation), each patient-derived Tat required specific primers for PCR amplification. The primers as follows:
19-c4 exon I
ATGGAGCCAGTAGATCCTAGACTAGAGCCCTGGAAGCATCCAGGAAGCCAGCCTAGGACTGCTTGT
ACCTCTTGCTATTGTAAAAAGTGTTGCTTTCATTGCCAAGTGTGTTTCATAACAAAAGGCTTAGGC
ATCTCCTATGGCAGGAAGAAGCGGGGACAGCGACGAAGAGCTCCTCAAGACAGTGAGACTCATCAA
GCGTCTCTATCAAAGCAG
19-c4-F1: 5′-GGGGTACCATGGAGCCAGTAGAT
19-c4-R1: 5′-GAGGTGGGCTGCTTTGATAGAG
19-c4 exon II
CCCACCTCCCAACCCCGAGGGGACCCGACAGGCCCGAAGGAACCGAAGAAGAAGGTGGAGAGAGAGA
CAGAGACAGATCCAGAACATTAG
19-c4-F2: 5′-TCAAAGCAGCCCACCTCCCAACC
19-c4-R2: 5′-CGGATATCCTAATGTTCTGGATC
The italicized nucleotides in the forward and reverse primers indicate the cleavage sites for the restriction endo nucleases KpnI and EcoRV, respectively, which correspond to the sequence necessary for directional cloning into the expression vector pcDNA3.1(+)/Hygro (Invitrogen, San Diego, CA), while the underlined portion is the respective start site of translation. This is termed 19-c4 Tat 101 throughout the manuscript.
Transient Expression Analyses
Jurkat or U-937 cells from exponentially growing cultures were seeded at 1 × 106 cells per 2 mL of growth medium and transfected with FuGene 6 transfection reagent in 35-mm 6-well plates. For each transfection, the following was added to each plasmid DNA preparation: firefly luciferase LTR expression constructs (LAI-LTR-luc or patient A19-derived LTRs or patient A19 clone18 mutants, 1000 ng) and 50 ngpRL-TK Renilla luciferase plasmid as an internal control vector DNA, premixed FuGene 6 (6 μL FuGene 6 with 94 μL of serum-free RPMI) was added drop wise. Co-transfection of Tat (500 ng) were performed with Tat IIIB 86 (Tat 86) or a full-length Tat derived from patient 107 (Tat 101). Empty vectors controls of the same amount were used as controls. After 30 minutes of incubation, the FuGene/DNA mixture was added drop wise to the cell cultures. After DNA delivery, transfected cells were incubated at 37°C for 24 hours, and cell extracts were harvested and assayed utilizing the dual-luciferase assay system as described by the manufacturer (Promega). Firefly luminescence was normalized to Renilla luminescence to control for variability in transfection efficiency. Firefly luminescence obtained with the pGL3 constructs was presented with the luminescence obtained with the parental HIV-1 LAI LTR activity set to 1.0 for each experiment and the relative activity of the mutagenized or the patient-derived LTR constructs was normalized relative to this value. Each experiment was performed in triplicate with a representative experiment shown. The error bars shown for each data point indicate the standard deviation from one representative experiment run in triplicate.
TNF κ Stimulation of Jurkat T Cells
After the transient transfection, transfected Jurkat cells were exposed to TNF-κ at a concentration of 10 ng/mL. Cells were exposed to TNF-κ for 24 hours and harvested for determination of HIV-1 LTR activity.
Statistical measures
For all transient transfection studies, confidence intervals were calculated using an alpha value of 0.1 (90%) and a data set of 9 as each experiment was performed in triplicate in three separate experiments. Significant differences (p< 0.05) were calculated using the Student T test for the comparisons as described and shown.
Patient 19-Derived LTR clones contain 5T Sp site III
HIV-infected patient 19 was identified as a patient who contained a 5T variant within Sp site III, which has been shown to be important in both the pre-HAART and HAART era as described above. Proviral DNA contained in peripheral blood mononuclear cells (PBMCs) from patient 19 was PCR amplified utilizing primers specific for the LTR; 24LTR/TA plasmid DNA clones for this patient were then obtained, as described in the Material and Methods. The LTR clones were sequenced (Genewiz, South Plainfield, NJ) to define the specific quasi species present in the peripheral blood compartment of patient 19. All LTRs were cloned from the PCR-amplified LTR product from patient 19 and their sequences were aligned to the LAI LTR laboratory strain using the MegAlign Program (DNASTAR, Madison, WI) (Figure 1). All of the LTR clones from patient 19 contain a 5T change at Sp site III. Based on the sequence analysis of the 24 LTR clones derived from patient 19, four were selected as representative clones for functional studies: clone 2 (19-2), clone 18 (19-18), clone 22 (19-22), and clone 24 (19-24) (Figure 1). Of the 24 clones obtained 18 had the same sequence as clone 2, 1 had the clone 18 sequence, 3 had the clone 22 sequence, and 2 had the clone 24 sequence.
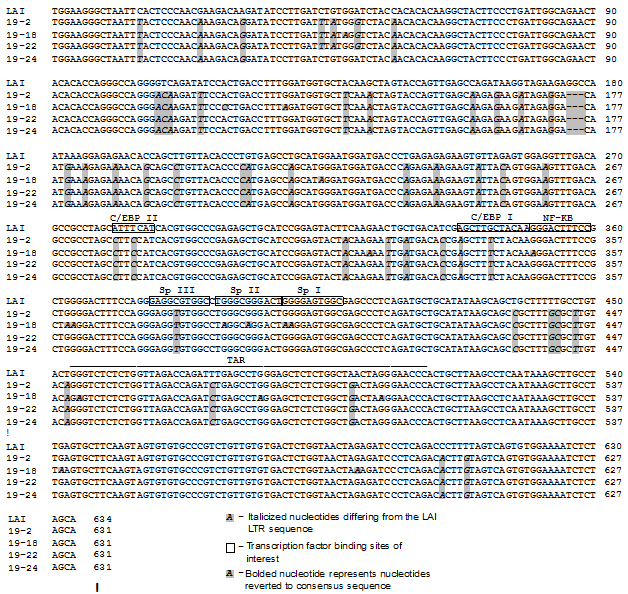
Figure 1 Patient 19 LTR clone sequences demonstrate differences throughout the LTR.
Four LTRs were cloned from patient 19 PCR-amplified LTR product and their sequences were aligned to the HIV-1 laboratory strain LAI LTR using the Meg Align Program (DNASTAR, Madison, WI). The clones derived from patient 19 and used in this study were labeled as clone 2 (19-2), clone 18 (19-18), clone 22 (19-22), and clone 24 (19-24).
To understand the functional properties of the patient 19–derived LTR variants, the LTR-driven pGL3 luciferase constructs were constructed from PCR products amplified from patient 19–derived LTR/TA clones. The sequences of the representative clones selected for analysis were aligned with the LAI LTR sequence (Figure 1). Each of the four LTR clones contained a number of nucleotide changes or insertions as compared with the LAI LTR sequence. Clones 19-2 and 19-22 have exactly the same sequence along the entire LTR. The clone 19-18 LTR exhibits the most nucleotide changes among the four clones compared with the consensus sequence. Their sequences were compared with the LAI LTR at eight transcription factor binding sites (C/EBP site II, ATF/CREB, C/EBP site I, NF-κB sites I and II, and Sp sites I, II, and III) as well as the TAR element. Clones 19-2, 19-22, and 19-24 have the same variation within the eight transcription factor binding sites and TAR element. The clone 19-18 LTR has nucleotide changes within NF-κB sites I and II, Sp sites I and II, and the TAR element (Figure 1).
Patient 19-Derived HIV-1 LTRs exhibit a different pattern of basal transcription activity within a t-cell line versus a monocytic cell line
To study the functional properties of the patient 19-derived LTR variants, or quasi species, the four LTR-driven luciferase constructs were transiently transfected into the Jurkat T-cell line in parallel with luciferase reporter constructs driven by the LAI LTR and the LAI LTR containing the 5T variant. The relative basal activity of each LTR clone is shown as the fold change over HIV-1 LAI LTR basal activity, which ranges from 0.5 with clone 19-18 to 1.5 with clone 19-24 (Figure 2A). Similar transient expression studies were also performed in the U-937 monocytic cell line (Figure 2A). In contrast to their activity in the T cell, a majority of the cloned patient-derived Sp site III5T-containing LTRs exhibited fold over HIV-1 LAI LTR basal activities below the activities obtained with the HIV-1 LAI LTR or LAI 5T-containing LTR, with clone 19-18 LTR exhibiting the lowest activity.
All Patient 19-Derived HIV-1 LTR clones containing a 5t sp site iii were responsive to hiv-1 tat86 except clone 19-18
The next question that was addressed was whether patient 19–derived LTRs could be transactivated by the transactivator protein Tat (derived from the widely used laboratory strain IIIB; Tat86 containing a naturally occurring deletion of 15 amino acids from the carboxyl terminus of exon II). To this end, co-transfection studies were performed with each LTR construct and the HIV-1 Tat86 expression vector in Jurkat T cells (Figure 2B). LTR clones 19-2, 19-22, and 19-24 were responsive to the HIV-1 Tat86, whereas the 19-18 LTR was defective with respect to Tat-mediated transactivation.
Co-transfections of each LTR construct with the HIV-1 Tat86 expression vector were also performed in U-937 cells (Figure 2C). Similar to the results observed in Jurkat T cells, all patient 19–derived LTRs, except 19-18 LTR, could be transactivated by HIV-1 Tat86. Though the Tat-stimulated transcription activities of 19-2 and 19-22 were significantly lower than that achieved with the Tat-stimulated LAI LTR when compared to the LAI basal level, the Tat-transactivated activities over each matched LTR basal level were the same or more to that obtained with the HIV-1 LAI LTR transactivated by Tat86 (Figure 2A). Furthermore, the clone 19-18 LTR was not transactivated by Tat 101, further suggesting that the defective nature of the 19-18 LTR was due to specific sequence alterations in the LTR (Figure 3).Furthermore, because the clone 19-18 LTR was nonresponsive to both Tat86 and Tat101, the defective nature of the LTR was not due to the absence of a protein domain in the naturally truncated version of Tat present in HIV-1 strain LAI.
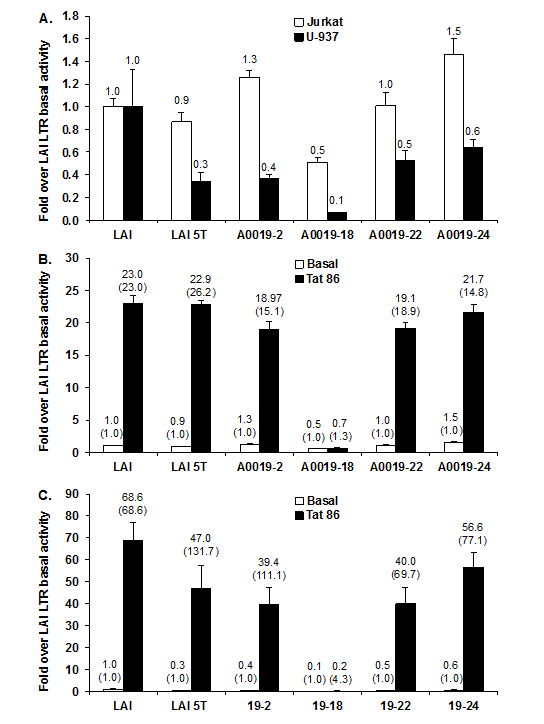
Figure 2 Patient 19 clone 18 LTR demonstrates lower basal transcription activity as compared with the HIV-1 laboratory strain LAI LTR and cannot be transactivated by the laboratory strain IIIB Tat 86 protein.
Patient-derived LTR luciferase reporter constructs (1000 ng) were transiently transfected into 1 × 106 Jurkat T cells/2 mL or 1 × 106 U-937 cells/2 mL using the Fu Gene 6 transfection reagent (Roche, Indianapolis, IN) in parallel with luciferase reporter constructs driven by the HIV-1 LAI LTR and the LAI 5T SpIII variant. LTR transcription activity was detected 24 hours after transfection using the dual-luciferase reporter assay.
Patient 19-derived LTR luciferase reporter constructs (1000 ng) and the HIV-1 Tat86 expression vector (500 ng) were co-transfected into Jurkat T cells (A and B) or U937 monocytic cells (A and C). LTR basal and Tat-mediated transcriptional activity was detected 24 hours after transfection using the dual-luciferase reporter assay. Results are presented as the mean values (±CI) over LAI basal activity and are representative of three transient expression assays performed in triplicate (9 total data points). The fold transactivation compared to its own basal is provided in parentheses.
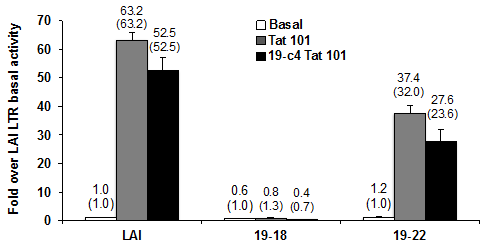
Figure 3 Patient clone 19-18 cannot be transactivated with either full-length or patient 19-derived full-length Tat.
Tat-mediated transcriptional activity were determined 24 hours after transfection using the dual-luciferase reporter assay and results are presented as the mean values (±CI) over LAI basal activity and are representative of three transient expression assays performed in triplicate (9 total data points). The fold transactivation compared to its own basal is provided in parentheses.
Patient 19–Derived tat protein could not transactivate the hiv-1 clone 19-18 ltr
To determine if Tat derived from the peripheral blood compartment of the same patient could activate the defective clone 19-18 LTR, we cloned full-length patient 19 clone 4 Tat (19-c4 Tat101) into the expression vector pcDNA3.1(+)/ Hygro. The sequence alignment of the patient 19 clone 4 (19-c4) Tat101 protein compared with the HIV-1 IIIB Tat and the consensus B (conB) Tat using Biology Work Bench 3.2 (data not shown) indicated that one conservative amino acid change of a strong group (K19R: lysine to arginine change at residue 19) and two conservative changes of weak groups reside within the transactivation domain of the Tat protein (residues 1-48).A couple of non consensus changes were shown to reside outside the transactivation domain (R53G: arginine to glycine change at residue 53, and F/V100E: phenylalanine or valine to glutamic acid change at residue 100).The 19-18 LTR or 19-22-LTR luciferase reporter constructs were transfected into Jurkat T cells, in parallel with the HIV-1 LAI LTR luciferase reporter construct. Expression vectors containing HIV-1 Tat101 or patient 19-derived Tat101 were also co-transfected into each of these cell populations to assess their response to Tat. The results indicate that the 19-18 LTR was not responsive to either Tat101 or 19-c4 Tat101 (Figure 3). The results also show that 19-c4-Tat101 exhibited a similar level of transactivation as compared with Tat101 with respect to both the LAI LTR and 19-22 LTR. However, with HIV-1 Tat101 and 19-c4 Tat transactivation, the 19-22 LTR exhibited lower Tat-mediated transcription activity than the LAI LTR (Figure 3).
The HIV-1-infected patient 19 clone 18 ltr is not responsive to tnf-astimulation or to tnf-a-enhanced hiv-1 tat 86activation
Tumor necrosis factor-κ (TNF-κ)has been reported to increase NF-kB activity in HIV-1-infected PBMCs, which has been shown to enhance HIV LTR basal transcription.30 To examine the impact of NF-κB on transcription driven by patient 19–derived LTRs, Jurkat T cells were stimulated with human recombinant TNF-α for 3 hours after transient transfection with the patient-derived LTRs. The results shown in Fig. 4 demonstrated that activation of the NF-κB pathway by stimulation with TNF-α resulted in approximately a 5-fold activation of all patient 19–derived LTRs except clone 19-18, which was almost a 3-fold activation. TNF-α increased the Tat-mediated LTR activity when compared with the activity of the same LTR by 229-fold with the 19-2 LTR and by 213-fold with the 19-24 LTR. Compared with LTR-directed transcription driven by Tat86 alone (Figure 2B), TNF-α dramatically enhanced Tat-mediated LTR activity in Jurkat cells. However, TNF-α slightly increased basal and Tat-mediated transcription activity driven by the 19-18 LTR but was dramatically reduced in comparison to the other clones, suggesting that sequence variation in the 19-18 LTR other than 5T in Sp site III could cause the defective transcription activity (Figure 4).
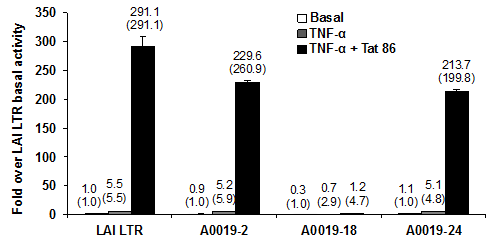
Figure 4 Patient 19 clone 18 LTR is slightly activated by TNF-α or by the viral transactivator protein Tat.
Patient-derived LTR luciferase reporter constructs and the HIV-1 Tat86 expression vector (500 ng) were co-transfected into Jurkat T cells. TNF-α was added 3 hours after transfection to the cell cultures at a final concentration of 10 ng/mL. LTR basal, TNF-α-activated, and Tat-mediated transcriptional activity was determined 24 hours after transfection using the dual-luciferase reporter assay and results are presented as the mean values (±CI) over LAI basal activity and are representative of three transient expression assays performed in triplicate (9 total data points). The fold transactivation compared to its own basal is provided in parentheses.
Reversion of the HIV-1 clone 19-18 ltr position +32 of tar to the lai base (tar32g) partially rescues tat-transactivated ltr activity
HIV-1 Tat functions through a unique complementary RNA regulatory segment of the LTR designated the TAR element (nucleotides +1 to +59 within the R region of the HIV-1 LTR), which forms a stable stem-bulge-loop structure.5 Sequence alignment of the patient 19 LTRs with the LAI LTR demonstrated that the clone 19-18 LTR had four nucleotide changes in the TAR element: 2A (G-to-A change at TAR position +2), 32A (G-to-A change at position +32), 52A (G-to-A change at position +52), and 47G (A-to-G change at position +47, which existed in all the other patient 19 LTR clones) (Figure 1). Analysis of the sequence variation within the TAR region of 19-18 LTR utilizing predictive RNA secondary structure software exhibited the bulge (+23 to +25) and the loop region (+30 to +35), which have been shown to be important for the binding and transactivation of Tat (Figure 5). The genetic variation present in clone 19-18 as compared with the LAI sequence predicted an alteration in the structure of the base of the stem that could theoretically account for the loss in Tat-mediated transactivation with this LTR clone. Based on this prediction, site-directed mutagenesis (SDM) was performed to convert A to G at TAR position +2, +32, and +52 of 19-18 LTR (the LAI nucleotide at these three positions), respectively. Subsequently, co-transfection studies were performed with each 19-18 LTR variant and the HIV-1 Tat101 expression vector in Jurkat T cells (Figure 6A).
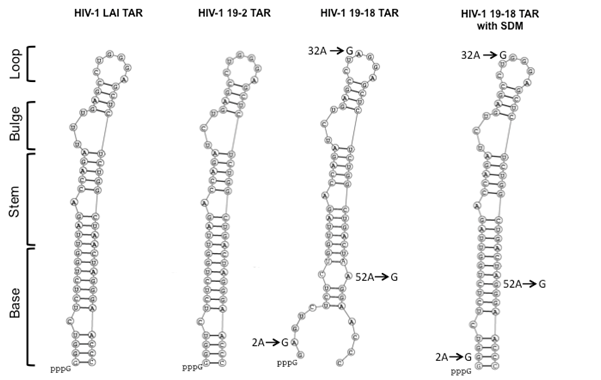
Figure 5 Mapping TAR variations in the LAI LTR versus the patient 19 clone 18 LTR.
Analysis of the sequence variation within the TAR region of patient 19 clone 18 utilizing predictive RNA secondary structure software depicts the bulge and loop regions, which are important for the binding and transactivation of Tat, respectively. Arrows show the nucleotide changes in the patient 19-18 LTR TAR element compared with the LAI TAR element.
As a positive control for patient 19–derived LTR clones, 19-22 LTR exhibited the same basal transcription activity as the LAI LTR but a lower Tat-mediated transcription activity than the LAI LTR. Similar to the results obtained with the original19-18 LTR, LTRs containing the LAI change at TAR position +2(TAR2G) and the LTR containing the LAI change at TAR position +52 (TAR52G) were not responsive to HIV-1 LAI Tat101.As predicted, the HIV-1 clone 19-18 LTR containing the LAI change at TAR position +32(TAR32G) partially rescued Tat transactivation of the clone 19-18 LTR. Furthermore, combinational clones of the LAI bases at the three positions were constructed and promoter activity studies in Jurkat T cells demonstrated that all the constructs containing TAR32G could only partially rescue Tat-mediated transcription activity of 19-18 LTR compared with Tat transactivation of 19-22 LTR (Figure 6B).
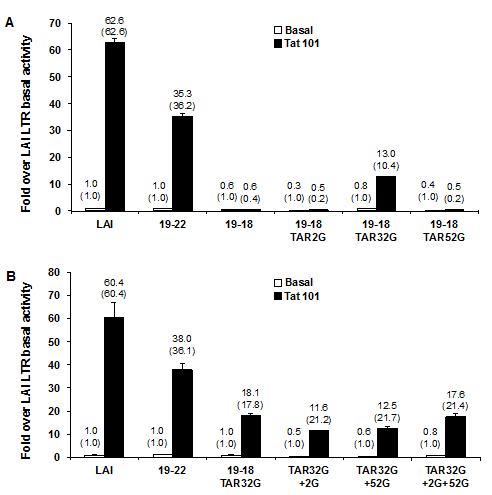
Figure 6 Reversion of patient 19 clone 18 position 32 of TAR to the LAI sequence (A to G) partially rescues Tat-transactivated LTR activity. SDM was used to convert positions 2, 32, and 52 within TAR of patient 19 clone 18 LTR luciferase reporter construct (19-18 LTR) to LAI genotypes (19-18 TAR2G, 19-18 TAR32G, 19-18 TAR52G).
The 19-18 LTR or the clone variants were transfected into Jurkat T cells with or without the HIV-1 Tat101 expression vector
Conversion of the hiv-1 19-18 ltr to tar32 lai with sp lai binding sites enhanced restoration of tat-mediated transactivation of the clone 19-18ltr containing the tar 32 (lai) sequence change
Sequence alterations within the Sp elements in the core region of the LTR have been shown to alter Tat-mediated LTR activation. Deletion of the LTR Sp binding sites dramatically reduces Tat transactivation.16 Sequence alignment of the patient 19 LTRs with the LAI LTR sequence demonstrated that the clone 19-18 LTR contained not only the 5T configuration of SpIII but also the 1A2A configuration (G-to-A change at positions 1 and 2 of Sp site I) and the 2A6A binding site II variant (G-to-A change at positions 2 and 6in Sp site II) (Figure 1). To determine whether the genetic variants in the Sp binding sites were also involved in the loss of Tat-mediated transactivation, each of the three Sp binding sites (SpI, SpII, and SpIII individually), or Sp sites I and II, or all Sp sites together were converted to the LAI configuration and transient co-transfections were performed in the absence or presence of HIV-1 Tat101 in Jurkat T cells. Although this finding was not altogether unexpected, none of the changes in the Sp binding sites resulted in any level of restoration of Tat-mediated transactivation of the modified clone 19-18 LTRs with Tat101 (Figure 7). These results also show that the 19-18 parental clone as well as SpI LAI, SpII LAI, SpIII LAI and All Sp LAI have similar reduced levels, regardless of the reverted Sp binding sites (Figure 7). Similar results for these reversions were seen in an experiment conducted in the U-937 monocytic cell line (data not shown).

Figure 7 Alteration of patient 19-18 LTR Sp binding sites alone does not alter the response to HIV-1 LAI Tat transactivation.
SDM was used to convert each or combinations of the three Sp binding sites (I, II, III) within patient 19 clone 18 LTR luciferase reporter construct (19-18 LTR) to LAI genotypes (SpI LAI; SpII LAI; SpIII LAI; SpI and II LAI; or all Sp, i.e., SpI, II, and III LAI). The 19-18 LTR of each variant was transfected into Jurkat T cells in the absence or presence of co-transfection with the HIV-1 Tat101 expression vector. LTR basal and Tat-mediated transcriptional activity was detected 24 hours after transfection using the dual-luciferase reporter assay and results are presented as mean values (±CI) over LAI basal activity and are representative of three transient expression assays performed in triplicate (9 total data points). The fold transactivation compared to its own basal is provided in parentheses. HIV-1 LAI basal (1.00 fold) and Tat-mediated transactivation (56.4 fold) was not shown as this would skew the y-axis to the point of not being able to see the results of the Sp site reversions.
However, when the Sp binding sites were converted to the LAI sequence configuration in the background of the 19-18 TAR32G LAI clone (TAR32G in conjunction with each individual Sp site or all three LAI Sp binding site configurations), transient expression assays demonstrated an enhanced restoration of Tat-mediated transactivation of 19-18-LTR TAR32G in both Jurkat and U-937 cells (Figure 8). The transient expression results obtained in Jurkat T cells also showed that the change over their own LTR basal activity values increased from 15.9-fold with the TAR32G to 22.4-fold with the TAR32G alteration in conjunction with Sp III converted to the Sp LAI sequence and to 32.4-fold with the TAR32G alteration in conjunction with all three Sp sites converted to the LAI sequence (values close to the Tat transactivation of the parental 19-22 LTR). Similar Tat transactivation results were obtained with the parental 19-22 LTR in U-937 cells. Increases over LAI basal activity were17.1-, 15.2-, and 50.5-fold with the TAR32G alteration; TAR32G alteration in conjunction with Sp III converted to the Sp LAI sequence; and TAR32G alteration in conjunction with all three Sp sites converted to the LAI sequence, respectively. However very interestingly, due to the lower basal level of the LTR, when you compare the fold increase to their own basal LTR activity all but TAR32G with SpI reach LAI transactivation levels or more. Cumulatively, these observations indicated that the nucleotide changes that resulted in the conversion of the binding sites to the LAI sequence configuration enhanced the response of the 19-18 LTR to Tat only in the presence of a more functional TAR element.
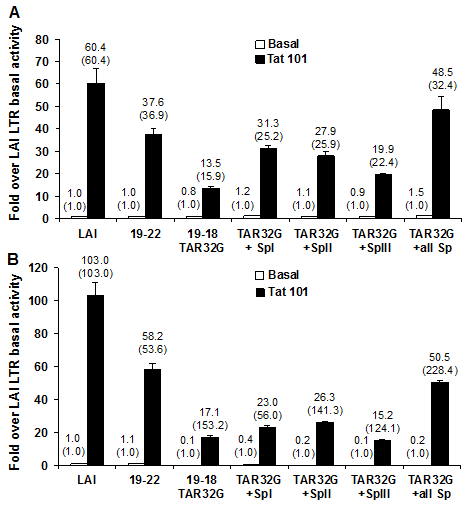
Figure 8 Reversion of patient 19 clone 18 position 32 of TAR to the LAI sequence (A to G) with conversion of Sp binding sites to LAI sequences enhances restoration of Tat-mediated transactivation of the 19-18 LTR. Based on 19-18-LTR TAR32, SDM was used to convert each individually or all of the three Sp binding sites (I, II, III) to LAI genotypes (TAR32 + SpI LAI, TAR32 + SpII LAI, TAR32 + SpIII LAI, or TAR32 + all Sp LAI). The 19-18-LTR TAR32 of each variant was transfected into 1 × 106 Jurkat T cells/2 mL (A) or 1 × 106 U-937 cells/2 mL (B) using the FuGene 6 transfection reagentin the absence or presence of co-transfection with the HIV-1 Tat101 expression vector. LTR basal and Tat-mediated transcriptional activity was detected 24 hours after transfection using the dual-luciferase reporter assay and results are presented as the mean values (±CI) over LAI basal activity and are representative of three transient expression assays performed in triplicate (9 total data points). The fold transactivation compared to its own basal is provided in parentheses.
We have reported here that three LTR clones (19-2, 19-22, and 19-24) derived from patient 19 have similar basal, stimulated, and Tat-mediated transcription when compared with the LAI LTR in Jurkat T cells when screening across a series of LTRs derived from more than 20 patients ((24)and unpublished data). However, one clone also derived from patient 19, now designated the clone 19-18 LTR, exhibited a marginally lower basal level of transcription in Jurkat T cells but much lower basal transcription in U-937 cells, suggesting that LTR-mediated gene expression may be modulated by different transcription factors in a cell type–dependent manner. This result is consistent with reports showing that different cells can affect the level of HIV-1 replication and gene expression owing to their distinct expression of cell type–specific transcription factors.31-33 We have also reported herein that the 19-18 HIV-1 LTR clone did not respond to Tat-mediated transactivation with strain IIIB Tat in Jurkat T cells and U-937 cells (Figure 2). Sequences located in both the trinucleotide bulge and the hexanucleotide G-rich loop of HIV-1 TAR have been shown to be required for Tat function.7,34 We hypothesized that the sequence alterations in the TAR region of 19-18 LTR could be the reason for the nonresponsive nature of the defective 19-18 LTR to Tat transactivation, especially the TAR32A mutation that is located in the loop. The LAI configuration TAR32G partially rescued the 19-18 LTR’s response to Tat 86 and 101. In contrast, neither of the LAI configurations (TAR2G or TAR52G) could reverse the defective nature of the 19-18 LTR with respect to Tat transactivation in either a representative T-cell or monocytic cell line (Figure 6).
It is widely accepted that binding of Tat to TAR is essential for Tat-mediated transactivation. HIV-1 Tat could bind to both wild-type or loop mutants of TAR but not to TAR containing alterations affecting the bulge region.35 Tat–TAR interactions involve the participation of several cellular co-factors. The activation domain of Tat is critical for recruiting p-TEFb, which is composed of cyclin T1 and CDK9, to TAR. Many studies have shown that Tat utilizes the transactivation domain to recruit p-TEFb to TAR; thus Tat could bind to the TAR bulge and p-TEFb to the TAR loop.36 The interaction of Tat with cyclin T1 and the TAR element results in hyper phosphorylation of the C-terminal domain by CDK9 and in subsequent increased processivity of RNA polymerase II.37 However, studies have also indicated that Tat could function in the absence of TAR and recruit cellular factors to the transcription complex.38 A cellular general transcription factor TFIIIH kinase (CAK) can also bind to the activation domain of Tat and phosphorylate the RNA pol II carboxyterminal domain. As part of the preinitiation transcription complex, TFIIH assembles on the promoter and then departs from the complex during the early elongation phase of transcription.39 Tat could stimulate both early and late phases of the transcription elongation by interacting with the two kinases CAK and CDK9. The results reported in this paper have demonstrated that HIV-1 TAR with the intact bulge-and-loop structure is required for both the full-length and truncated Tat-mediated LTR-driven transcription.
The Sp binding sites are important in Tat-mediated transcription.40 The core region of the LTR is composed of the TATAA box, which is located on 29 to 24 bases upstream of the transcriptional start site, and three tandem GC-rich binding sites (-45 to -77) shown to bind members of the Sp family of transcription factors. Functional analyses of HIV-1 LTR-driven reporter constructs have shown that mutations with all three Sp elements dramatically decreased both basal and Tat-mediated transcriptional activation in HeLa cells.16 Moreover, studies using HIV-1 molecular clone constructs have shown that removal of the GC box array leads to a reduction in viral replication in most, but not all, cell types.41-44 However, mutations of one or two of the three Sp sites have little effect on basal or Tat-driven transcription. In addition, the mutation of the two NF-κB motifs and all three Sp binding sites within HIV-1 LTR led to defective viral replication.41-45 Herein, the results have shown that HIV-1-infected patient–derived LTR clone 19-18 contains nucleotide changes within all three Sp sites compared with the LAI sequences (1A2A in Sp site I, 2A6A in SpII, and 5T in SpIII; Figure 1). Similar to previous results reported in HeLa cells,16 the sequence alterations with all three Sp elements within 19-18 LTR resulted in reduced basal activity in Jurkat T cells and even further reduction in U-937 cells (Figure 2, 3A, 4A, 7, and 8). In addition, the mutations within all three Sp elements within the functional TAR-containing LTR 19-18 TAR32G exhibited significantly lower Tat-mediated transcription activity than the HIV-1 LAI LTR or other patient 19–derived LTR clones in Jurkat T cells and U-937 cells, which is consistent with results reported previously.42-44
Sp transcription factors regulate expression of a number of viral genes via interactions with gene promoters as well as other regulatory proteins. For example, Sp factors binding to the GC-rich region of the HIV-1 LTR may facilitate Tat recruitment proximal to the basal transcriptional machinery.46 SpI is able to interact with either Vpr or C/EBPβ to cooperatively activate HIV-1 gene transcription in a cell type–specific manner.47,48 One study has revealed that HIV-1 Tat could contact both Sp1 and DNA protein kinase, thus enhancing the DNA-protein kinase–mediated phosphorylation of Sp1.49 An earlier investigation also indicated a direct physical interaction between Sp1 and Tat.50 However, a recent study using the yeast two-hybrid assay showed that Sp1 does not interact directly with Tat.51 In our studies, the nucleotide changes that resulted in the conversion of the three Sp binding sites to the LAI sequence configuration enhanced the response of the 19-18 LTR to Tat only in the presence of a functional TAR element containing the intact loop structure (Figures 7 & 8). These results suggested functional connectivity between Sp factor binding to the GC box array and a functional TAR with regard to Tat-mediated transactivation.
The fact that the 19-18 LTR transcription activity could not be enhanced by TNF-κ stimulation of Jurkat T cells (Figure 4) may be explained by the sequence alterations located in the NF-kB sites I and II (1A2A NF-kB site I and 1A NF-κB site II). Cytoκines play a critical role in the pathogenesis of HIV-1. Interleuκin-6, TNF-α, interleuκin-1β, and other pro inflammatory cytoκine levels are elevated in the blood, bone marrow, and cerebrospinal fluid of HIV-1-infected patients.52 Interleuκin-6 and TNF-α are induced early after HIV monocytic infection, followed by their continued increased expression. TNF-α is one of the most potent activators of NF-κB activity κnown. It acts by inducing a signaling cascade that activates the IκB κinase complex, which phosphorylates IκB, releasing NF-κB. The free NF-κB translocates to the nucleus and induces the activation of the HIV-1 LTR. TNF-α has been reported to increase NF-κB activity in HIV-1-infected PBMCs, which enhances HIV-1 LTR activity driven by Tat.30 The functional studies reported in this paper have shown that TNF-α can enhance the basal and Tat-mediated transcription activity driven by patient 19–derived LTRs containing the consensus NF-κB site I and II. Specific sequence alterations in NF-κB site I and II could lead to inability of TNF-α to stimulate basal and Tat-mediated transcriptional activity.
In summary, these results have demonstrated that the defective patient-derived LTR 19-18 is nonresponsive to Tat transactivation, primarily owing to a single-base pair alteration in the loop structure of TAR at position 32. Conversion of Sp binding sites to LAI sequences enhanced restoration of Tat-mediated transactivation of the 19-18 LTR with a functional TAR region, suggesting that SpI factors facilitate Tat transactivation. Cumulatively, these experimental observations suggest that in some cases relatively severe defects in Tat-mediated transactivation result from a relatively small amount of sequence variation within the viral promoter. In these cases, one might expect that a small amount of gene transcription from the defective provirus might lead, over a prolonged HIV disease course, to additional genetic variation that could result in the recovery of viral fitness in at least a subset of viral genomes. In addition, it may also be expected that the Tat directly linked to LTRs with TAR defects may also have alterations that could either complement the ones observed in the LTR recovering some if not all of the function or may make the viral genome even more inefficient and not able to replicate.
These studies were funded in part by the Public Health Service, National Institutes of Health, through grants from the National Institute of Neurological Disorders and Stroke, NS32092 and NS46263, the National Institute of Drug Abuse, DA19807 (Dr. Brian Wigdahl, Principal Investigator), National Institute of Mental Health Comprehensive Neuro AIDS Core Center (CNAC), P30 MH-092177 (KamelKhalili, PI; Brian Wigdahl, PI of the Drexel subcontract), and under the Ruth L. Kirschstein National Research Service Award 5T32MH079785 (Jay Rappaport, PI, Brian Wigdahl, PI of the Drexel subcontract). The contents of the paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Dr. Michael Nonnemacher was also supported by faculty development funds provided by the Department of Microbiology and Immunology and the Institute for Molecular Medicine and Infectious Disease.
Approval: Patients in the Drexel Medicine CARES Cohort were recruited under protocol 16311 approved by the Drexel University College of Medicine IRB, which adheres to the ethical standards of the Helsinki Declaration (1964, amended most recently in 2008), which was developed by the World Medical Association as described. Consent for enrollment: All patients provided written consent upon enrollment. Consent to publish: The consent form signed at each visit by each patient contains approval from each patient for the publication of results obtained during their clinical visits and thereafter during experimentation performed with clinical material derived during their visits.
None.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
 World Malaria Day, observed annually on April 25SUP>th. The purpose of this day is to raise awareness about the global struggle to combat malaria, highlighting the need for continued research, prevention, and treatment strategies to eradicate this deadly disease. So, it is an opportunity to all the researchers to submit your interesting papers on raising the approachability on modern vaccine development, advances in disease control, prevention and eradication and the submissions received till April 25th, 2024 will be offered a best discount of 30% for publication in Journal of Human Virology & Retro virology.
World Malaria Day, observed annually on April 25SUP>th. The purpose of this day is to raise awareness about the global struggle to combat malaria, highlighting the need for continued research, prevention, and treatment strategies to eradicate this deadly disease. So, it is an opportunity to all the researchers to submit your interesting papers on raising the approachability on modern vaccine development, advances in disease control, prevention and eradication and the submissions received till April 25th, 2024 will be offered a best discount of 30% for publication in Journal of Human Virology & Retro virology.