Journal of
eISSN: 2373-6453


With the enhancement of high-throughput screening (HTS) for target-based antiretroviral discovery, fluorescence has been considered the best bioassay. Bioluminescence by genetic reporters has maintained a major place among cell-based virological assay formats. Luminescent bioassays have a wide variety of applications due to their high sensitivity and linearity, even for a range of complex biological samples. In this target-based method, we used an HTS bioluminescence assay and highlighted the detection capabilities of this time-of-addition experiment.
Hence, the development of a cell-based assay that uses a phenotypic drug discovery approach based on bioluminescence by stable reporter cells is described. Utilizing this screening method, a bioluminescent, target-based, time-of-addition experiment was performed by adding antiretrovirals with known mechanisms of action to analyze their drug targets by measuring the length of time until the antiretrovirals lost their efficacy in an HIV-1 replication assay.
Depending on the viral replication target, the antiretrovirals lost activity at different times: fusion inhibitors acted for 0-2 h; retrotranscriptase inhibitors acted efficiently for the first 4 h; integrase inhibitors acted for 18 h; and protease inhibitors were inactivated after 15 h.
The target-based, bioluminescence assay identifies the mode of action of antiretroviral drugs and provides valuable information about drug targets that inhibit HIV replication. This assay delimits the time (hours) for which the addition of an antiviral can be delayed before losing its antiviral activity relative to the replication cycle of HIV. The target of an antiviral compound can be identified both by comparing the relative time until it loses efficacy to that of the reference drugs and by the time elapsed when the antiretroviral loses activity compared to a known control.
Our target-based bioluminescence assay is fast, reliable, sensitive and useful for high-throughput drug screening.
Keywords:HIV, Time-of-addition experiment, Antiretroviral, Bioluminescence assay, Fusion inhibitor, High-throughput screening, Reporter gene, Indicator cell
HAART, Highly Active Antiretroviral Therapy; HTS, High-throughput Screening Assay; FI, Fusion Inhibitors; NRTI, Nucleoside Reverse Transcriptase Inhibitor; NNRTI, Non-Nucleoside Reverse Transcriptase Inhibitor; INI, Integrase Inhibitor; PI, Protease Inhibitor; MI, Maturation Inhibitor; ARV, Antiretroviral; HIV-1, Human Immunodeficiency Virus; PCR, Polymerase Chain Reaction; TAE, Time-of-Addition Experiment; β-Gal, β- Galactosidase; MAGI, Multinuclear-Activation Galactosidase Indicator Assay; LTR, Long Terminal Repeat; GFP, Green Fluorescent Protein; TAE-luc, Time-of-Addition Experiment based on Luciferase
Globally, according to UNAIDS, an estimated 35.3 (32.2–38.8) million people were living with HIV in 2012. HIV is now a treatable infection, and patients receiving medical treatment have a better prognosis than in the past due to the availability of highly active antiretroviral therapy (HAART).1,2 Unfortunately, the infection is still not curable or preventable. The side effects and toxicity of HAART as well as the emergence of drug resistance add to the urgency to continue to search for new strategies, interventions and methodologies as well as to develop more sensitive and easy-to-interpret, high-throughput screening (HTS), target-based assays in the search of new antiretrovirals (ARVs) with different mechanisms of action against the virus.3-5
An understanding of the HIV replication cycle is essential to design effective antiviral drugs with specific mechanisms of action. The replication cycle of human immunodeficiency virus type 1 (HIV-1) is a complicated, multistep process that depends on both viral and host cell factors.6-8
Therapies based on inhibiting HIV replication initially targeted only the viral enzymes, which are exclusively expressed by the virus and are not present in the uninfected human host cell. HIV encodes three enzymes required for replication: HIV-1 reverse transcriptase, HIV-integrase and HIV-protease.9
Therefore, anti-HIV ARVs are classified into six different classes based mainly on their molecular mechanism of action: Entry/Fusion inhibitors (FIs), Nucleoside reverse transcriptase inhibitors (NRTIs), Non-Nucleoside reverse transcriptase inhibitors (NNRTIs), Protease inhibitors (PIs), Integrase inhibitors (INIs), and Maturation inhibitors (MIs).10
Deciphering the molecular target of an anti-HIV drug is considered an important tool for understanding the mechanism of action of the virus itself and gives us new insight into the various therapeutic approaches for clinical treatment, discovery of new ARVs and refinement of ARV drug design and development. There have been several studies on the mechanism of action of newly discovered HIV inhibitors, including virus-based assays, structure-based drug design, receptor pharmacology, biochemical screening, time-of-addition experiments and bioluminescence assays.11-13
Screening assays
Screening and testing the target of a novel ARV are important steps in antiviral innovation. Recently, screening assays have become essential for early-phase ARV discovery because of the advancement in drug synthesis technologies and because of a considerable increase in new pharmacologic targets.11,14,15
Most methods are intended either to identify the virus itself or to identify a specific antiviral target at a stage of the viral cycle using either flow cytometry, electron microscopy, qualitative PCR, fluorescent quantitative real-time PCR (FQ-PCR), or enzyme-linked immunosorbent assays (ELISA).16-20 Nevertheless, other assays elucidate the target of action of antiviral drugs in vitro. For example, by comparing the viral inhibitory activity of an antiviral agent against persistently or latently infected cells, it can be easily determined whether an ARV drug targets a pre- or post-integration step of the viral replication cycle in the host cell. An antiviral blocking the production of new virus from persistently infected cells suggests that the ARV most likely targets a post-integration event because, during persistent infection, no pre-integration steps, such as virus binding, reverse transcription and integration, are observed. For other steps in the HIV replication cycle (e.g. virus binding, integration, transcription and viral budding), specific cellular assays can be performed to investigate the outcome of a promising inhibitor of a specific target. For example, it can be hypothesized that if an ARV blocks viral binding, a specific virus-binding assay can be implemented. Nevertheless, none of the above-mentioned methods combine all of these properties into one assay that can assess the target alongside the viral replication cycle. This type of assay is termed a Time-of-Addition Experiment (TAE) because it tests the inhibition of ARVs at different point-times to elucidate the target of a drug.
Time-of-addition experiment
The search for and development of new ARV drugs that can lead to rapid effects on HIV requires a screening method that allows rapid determination of the antiviral mechanism of action.21,22 Screening target-based bioassays to elucidate the mechanism of action of new ARVs is of great significance to increase the development speed of novel anti-HIV ARVs and are a requirement for clinical development.23
A screening method to delimit the stages of the entire viral life cycle is a TAE.23 This time-based screening bioassay can determine, over the entire cell cycle, which stage of viral replication is being targeted by an antiviral drug.24,25
TAEs are cell-based assays that utilize the multinuclear-activation galactosidase indicator assay (MAGI). In this assay, infection results in the expression of a stable, integrated bacterial β-galactosidase (β-Gal) gene under the transcriptional control of an HIV-1 long terminal repeat (LTR). The expressed cellular β-Gal enzyme can be detected colorimetrically (i.e. by the detection of blue-colored cells, counting syncytial formation, measuring the viral protein p24 levels or using green fluorescent protein (GFP)26,27)). However, these assays are not as sensitive as the luciferase reaction.
Bioluminescence is currently a good option as a target-based method for high-throughput screening (HTS)28 because of its speed and, more importantly, its high sensitivity in detecting the viral protein and its capability to quantify small sample volumes.16,29 This target-based bioluminescence assay for HTS using β-galactosidase (β-Gal) was performed by our group using a HeLa-CD4-LTR-β-gal stable cell line and was easy to interpret, rapid and sensitive.
TAE-Luc on stages of the HIV cell cycle
Luciferase-based assays have a wide range of applications due to their high sensitivity, broad linearity, and robustness toward a variety of drugs and complex biological samples. Moreover, as the assay is based upon light emission activated by a Tat-dependent promoter, the assay can be easily used for HTS of ARVs.30
The need for a sensitive, HTS, time-based bioassay that is easy to interpret, rapid, reliable, and inexpensive for drug screening and can be validated using known ARVs21 with specific mechanisms of action led us to develop this protocol using the current technology of a luciferase bioassay.
This bioassay is based on the time of addition of ARVs and uses a β-Gal measurement output to quantify HIV infectivity. This protocol will help virologists and ARV developers to easily screen novel medications to elucidate their mechanisms of action against HIV.
Our Time-of-Addition Experiment based on Luciferase (TAE-luc) has an advantage over other assays in that a single experiment will give an indication of the target of an inhibitor and provide a basis for further investigations. However, for many viruses, at least a few inhibitors with an established mode of action are already known. A limitation of screening using HTS TAE-luc is the lack of well-characterized inhibitors for all viruses of medical interest and the need for indicator cells for other viruses. The use of well-characterized viral inhibitors that can be used as controls for specific viruses will improve the precision with which TAE-luc can reveal the target of action of a new antiviral inhibitor. A second limiting factor is that results obtained from some compounds with limited selectivity are not easily interpretable. Indeed, the use of compound concentrations of 10- to 100-fold their antiviral activity (half-maximum inhibitory concentration or IC50) are most favorable in this experiment, and compounds with limited selectivity can often be toxic to the target cells at these high concentrations.23,25,31-33
The percentage of residual infectivity after treatment was calculated with respect to the positive control (untreated virus) for each time period. This bioassay is extremely sensitive and specific to HIV-Tat, making it suitable for HTS with TAE. Here, we describe a standardized method that is easy to interpret and delimits time well. We determined the time needed for effective drug activity at each stage of the HIV replication cycle and found the time point at which the drug should be added within the HIV replication cycle for efficacy.
Reagents
Stock solutions of the ARVs were prepared in RPMI 1640 cell culture media (Invitrogen, Carlsbad, CA, USA). Subsequent serial dilutions of the stock were made in the culture media.
Cells, HIV-1 isolates, and antiretrovirals
HeLa-CD4-LTR-β-gal cells were provided by Dr. Michael Emerman (Harrington, Wu, Pullen, and Emerman, 2000b). HIV-1IIIB, Zidovudine or AZT (NRTI), indinavir (PI), 118-D-24 (INI), amprenavir (PI), and enfuvirtide or T-20 (FI) were provided by Dr. Suzanne Gartner, Dr. Mikulas Popovic and Dr. Robert Gallo and were obtained through the AIDS Research and Reference Reagent Program, NIH. Bacitracin (Anti-Protein Disulfide Isomerase) was obtained from Sigma-Aldrich, and UC781 (NNRTI) was kindly donated by Gadi Borkow.
Bioluminescence time-of-addition experiment (TAE-luc)
The compounds were added at the time of HIV-1 inoculation (time zero) or at various time points post-inoculation (0, 2, 4, 6, 8, 10, 15, 18, 24 h) to delimit the different ARV target stages of the viral cycle (fusion or entry, retrotranscription, protease activity, and integration into the genome). The antiviral compounds were added at a concentration several times their EC50, which is the concentration required to reduce the cytopathicity of HIV-1IIIB by 50%. Infection inhibition was quantified after 24 h by measuring the β-gal luciferase activity of the cells with the Beta-Glo Assay System (Promega Kit) using the Glo-Max Luminometer and software from Promega.
Statistical analysis
The data were graphed with the SigmaPlot 10.0 software, and the values shown are the means ± standard deviations of three separate experiments. Each experiment was performed in duplicate.
Time (site) of intervention
To determine the elapsed time (hours) for each specific target during the HIV replication cycle, TAE-luc was performed using different ARVs with known specific mechanisms of action. In the first set of experiments, HeLa cells (expressing CD4, CXCR4 and CCR5) were infected with HIV-1IIIB cell-free virus, and different ARVs were added to the infected cells at nine time intervals during the replication cycle of the virus (0, 2, 4, 6, 8, 10, 15, 18, 24 h).
The time-based addition of ARVs following the initiation of infection effectively inhibited virus replication, with a decreasing impact of further ARV addition over time that was dependent on the target and mechanism of action.
The TAE-luc assay was performed on the HIV replication cycle using indicator cells. The chosen ARVs targeted the following specific stages of the viral cycle: a) fusion or entry (T20, bacitracin), b) retrotranscription (UC-781, AZT), c) protease activity (amprenavir, indinavir), or d) integration into the cell host genome (118-D-24). As observed (Figure 1), T20 and bacitracin showed inhibition from 0 h to 2 h. The reverse transcriptase inhibitors UC781 and AZT efficiently inhibited the replication of HIV for 4 h (Figure 2). The PIs amprenavir and indinavir inhibited HIV activity at constant levels for up to 15 h; after this time, they began to lose inhibition activity (Figure 3). The INI 118-D-24 began losing anti-HIV activity after 18 h, as expected (Figure 4).
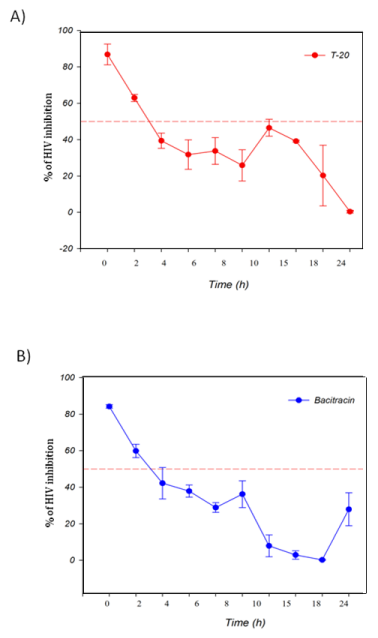
Figure 1 Time of intervention in HIV-1 life cycle for fusion inhibitors, HeLa/CD4-LTR-β-gal cells were infected with HIV-1IIIB cell-free virus before A) T-20 (100 μM), B) Bacitracin (3.5 mM) were added upon HIV-1 inoculation (time zero) or at various time points post-inoculation and β-gal activity was measured following 24 hr of incubation. Percentage values are relative to the positive control (no treatment). The data represent the means ± standard deviations from three separate experiments, each of which was carried out in duplicate.

Figure 2 Time of intervention in HIV-1 life cycle for reverse transcriptase inhibitors, HeLa/CD4-LTR-β-gal cells were infected with HIV-1IIIB cell-free virus before A) UC781 (70 nM), B) AZT (20.0 μM) were added upon HIV-1 inoculation (time zero) or at various time points post-inoculation and β-gal activity was measured following 24 hr of incubation. Percentage values are relative to the positive control (no treatment). The data represent the means ± standard deviations from three separate experiments, each of which was carried out in duplicate.
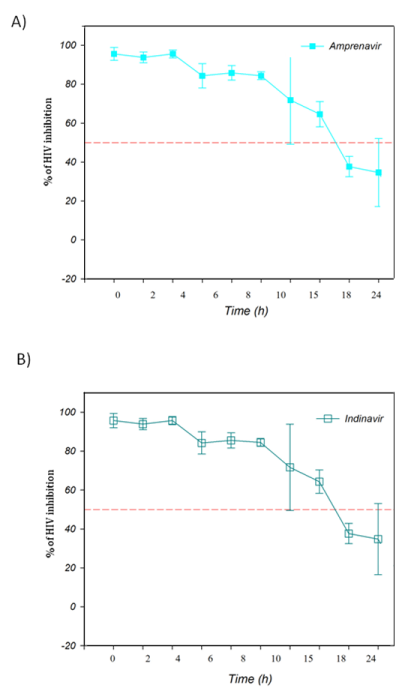
Figure 3 Time of intervention in HIV-1 life cycle for protease inhibitors, HeLa/CD4-LTR-β-gal cells were infected with HIV-1IIIB cell-free virus before A) Amprenavir (0.1 mM), B) Indinavir (0.25 μM) were added upon HIV-1 inoculation (time zero) or at various time points post-inoculation and β-gal activity was measured following 24 hr of incubation. Percentage values are relative to the positive control (no treatment). The data represent the means ± standard deviations from three separate experiments, each of which was carried out in duplicate.
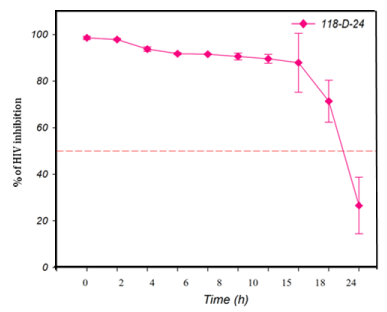
Figure 4 Time of intervention in HIV-1 life cycle for integrase inhibitors, HeLa/CD4-LTR-β-gal cells were infected with HIV-1IIIB cell-free virus before 118-D-24 (100 μM) was added upon HIV-1 inoculation (time zero) or at various time points post-inoculation and β-gal activity was measured following 24 hr of incubation. Percentage values are relative to the positive control (no treatment). The data represent the means ± standard deviations from three separate experiments, each of which was carried out in duplicate.
These results show that TAE-luc is a useful tool to delimit the stages at which HIV is inhibited by ARVs thus helping to determine the viral-replication-cycle targets of novel ARVs in host cells.
Both target-based and cell-based methods have been used to identify antiretroviral inhibitors of HIV. The target-based method helps to screen and identify antiretrovirals with identified mechanisms of action. Using this approach, we have identified inhibitors of the following mechanisms: a) fusion or entry (T20, bacitracin), b) retrotranscription (UC-781, AZT), c) protease activity (amprenavir, indinavir), and d) integration into the cell host genome (118-D-24). The cell-based method utilizes viral replication (e.g. replicon or complete virus infection) to screen for antiretrovirals that target either viral or host factors that are required for viral replication. This target- and luciferase-based, high-throughput screening method is a useful tool for the identification of HIV-life-cycle targets of inhibitors and is sensitive, reliable, cheap and fast. Tae-luc is simple to interpret, and various compounds can be measured at once. Moreover, results can be obtained in less than 2 days, making it a rapid and reproducible assay.
Our results show that entry/fusion occurs during the first 2 h of viral replication. After 2 h, FIs lose activity (Figure 1). It has been reported that during the first 4 h, reverse transcriptase inhibitors efficiently block the production of the RT enzyme. Integrase inhibitors act for up to 18 h, and protease inhibitors are effective for 15 h, which is nearly the complete replication cycle. Such results were similarly obtained in several other studies.11,34-37
Our TAE-luc has the advantage of being a sensitive, high-throughput, target-based screening bioluminescence assay that is faster than counting blue cells27 or syncytia formations,38,39 which are also less sensitive assays. As the HeLa cells are attached to the bottom of the well, it is easier to handle them, in contrast to the GFP cells,40,41 which are unattached or loose in the media. By adding the luciferase reagent, virus infection can be immediately quantified. There is also a luciferase-based method that involves the TZM-bl cell indicator;42 however, in our hands, we obtained a high background when working with these cells, and the group of Kim et al.43 had the same results. Thus, we recommend using the HeLa-CD4-LTR-β-gal cells, which have a lower background, resulting in a higher sensitivity.
With the knowledge of the time that passes until a certain drug loses efficacy by using TAE-luc, it will be easy to interpret the results of ARV mechanism-of-action studies. This is a sensitive, specific, rapid, inexpensive, and easily interpretable protocol. Therefore, this luciferase-based method is an option for the discovery of new targets of ARVs.
TAE-luc is a sensitive, reliable, fast and easy-to-interpret option as an HTS assay for ARVs and can be used as a screening method to identify the target of an inhibitor of the HIV replication cycle. This assay was performed using the known mechanisms of most of the known antiretroviral drugs and is easy to interpret because it measures the time to HIV inhibition in hours. The luciferase reagent can be added the day after the last drug was added.
None.
None.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
 World Malaria Day, observed annually on April 25SUP>th. The purpose of this day is to raise awareness about the global struggle to combat malaria, highlighting the need for continued research, prevention, and treatment strategies to eradicate this deadly disease. So, it is an opportunity to all the researchers to submit your interesting papers on raising the approachability on modern vaccine development, advances in disease control, prevention and eradication and the submissions received till April 25th, 2024 will be offered a best discount of 30% for publication in Journal of Human Virology & Retro virology.
World Malaria Day, observed annually on April 25SUP>th. The purpose of this day is to raise awareness about the global struggle to combat malaria, highlighting the need for continued research, prevention, and treatment strategies to eradicate this deadly disease. So, it is an opportunity to all the researchers to submit your interesting papers on raising the approachability on modern vaccine development, advances in disease control, prevention and eradication and the submissions received till April 25th, 2024 will be offered a best discount of 30% for publication in Journal of Human Virology & Retro virology.