Advances in
eISSN: 2373-6402


Research Article Volume 7 Issue 3
1Central Horticultural Experimental Station, India
2ivision of Crop Protection, Indian Vegetable Research Institute, India
3Indian Institute of Horticultural Research, India
4Department of Plant Pathology, University of Agricultural Sciences, India
Correspondence: V Venkataravanappa, Scientist (Plant Pathology) Division of Plant pathology Central Horticultural Experimental Station, ICAR-Indian Institute of Horticultural Research Chettalli- 571248, Kodagu, Karnataka, India
Received: October 20, 2016 | Published: July 6, 2017
Citation: : Venkataravanappa V, Reddy CNL, Manjunath M, et al. Detection, characterization and in-silico analysis of candidatus phytoplasma australasia associated with phyllody disease of sesame. Adv Plants Agric Res. 2017;7(3):288-300. DOI: 10.15406/apar.2017.07.00256
Leaf samples from sesame plants exhibiting Phyllody disease were collected from Varanasi and Mirzapur districts of Uttar Pradesh, India during the survey conducted between month of September to December, 2012-14. Incidence of sesame Phyllody in the farmers at different location was ranged from 30-70 percent indicating its prevalence in Uttar Pradesh. The Phytoplasma infection in sesame plants was confirmed by PCR using universal primers of 16s rRNA (R16F2n/R16R2) and SecY gene (SecYF2 and SecYR1) respectively. Amplified 16s rRNA and SecY gene was sequenced and sequence comparisons were made with the available Phytoplasma 16srRNA and SecY gene sequences in NCBI Gen Bank database. The 16srRNA and SecY gene sequence of Phytoplasma in the current study, shared highest nucleotide identity of 97.9-99.9% and 95.8 to 96.3% with subgroup 16Sr II-D the peanut witches’-broom group. A Comprehensive recombination analysis using RDP4 showed the evidence of inter- recombination in F2nR2 and SecY gene fragment of Phytoplasma infecting sesame. The most of the F2nR2 fragment is descended from Ash yellows-[16SrVIII] and Apple proliferation-[16SrX] group. While for SecY gene, most of the part was descended from Peanut witches'-broom- 16SrII-A (GU004331) and aster yellows 16Sr I-A (GU004345). The genetic similarities and the potential threat of this new Phytoplasma belong to 16Sr II-D subgroup of Peanut witches’ broom’ group infecting to sesame in north India are discussed.
Keywords: sesame phyllody, PCR, phytoplasma, 16SII group
Sesame (Sesamum indicum L.) is one of the most important and ancient oilseed crop grown in India and many parts of the world. Bulk of the world production of sesame is coming from Myanmar, India and China.1 In India, sesame is cultivated in an area of 1.83m ha with production of 0 .757m tonnes and productivity 413.6kg/ha. The productivity is low in India compared to world’s average (464.6kg/ha) and it is far below as compared to Egypt (1200kg/ha) and China (897.7kg/ha).2 Due to its high quality and quantity of oil (53.3%) and protein (25%), it is aptly called as the ‘queen’ of oilseeds.3,4 Sesame seed oil contains antioxidant sesame responsible for its long shelf life and oleic acid.3,4–7 Seeds and oil are used in cooking, salad, margarine and is also used as a raw material for the production of industrial products like insecticides, pharmaceuticals, paints, perfumes, soaps and varnishes.8,9 Sesame is vulnerable to biotic and abiotic stresses resulting considerable yield loss. In India, among biotic stresses, sesame Phyllody, is the most important disease appears in severe form affecting the plants partially or completely and having potential to cause yield loss upto100 per cent.10–12 Typical symptoms of this disease include floral virescens, Phyllody and proliferation of auxiliary shoots. However, sometimes these symptoms are found to be accompanied with yellowing, cracking of seed capsule, germination of seeds in the capsules and dark exudation on the foliage.13 Phytoplasma are phloem inhabiting, wall-less, obligate bacteria belonged to the class Mollicutes of prokaryotes.14,15 These sieve inhabiting pathogens spread in nature by sap sucking leaf hopper viz. Orosius orientalis, Circulifer haematoceps and Neoaliturus haematoceps in persistent manner.12,13,16–20 Phytoplsms are known to infect more than 1000 plant species including many agriculturally important crop species viz. fruits, vegetables, cereals, trees and legumes across the world.2,21–25 In the past, they were poorly understood because of their obligate nature and difficulty in culturing in vitro.26 The utilization of DNA-based methods for detection, characterization and phylogenic grouping based on highly conserved 16S rRNA gene among Phytoplasma provided better understanding of their diversity across the globe.27,28 Species specific and group specific primers to amplify 16SrRNA conserved gene in Phytoplasma were extensively exploited for the detection, identification as well as phylogenetic analysis.14,29,30 Based on the analysis of 16SrRNA sequences, 31 groups and 100 subgroups of diverse Phytoplasma were identified.31 These belong to 16SrI, 16SrII, 16SrV, 16SrVI, 16SrIX, 16SrXI and 16SrXIV groups. Among these, Aster yellows group (16SrI) is alone associated with more than 31 diseases and are reported from north-eastern parts of the country.32 So far, only few Phytoplasma diseases were reported from Eastern, Western and Central parts of India. Classification of distinct Phytoplasma strains below the species level has been based primarily on RFLP analysis of 16S rRNA gene sequences. Epidemiological studies of diverse Phytoplasma strains over a period, which are very closely related based on analysis of 16SrRNA gene sequences known to be associated with similar diseases in different cultivars of a given plant species grown in the same or different geographical regions.33–36 Often, such strains cannot be readily differentiated by analysis of 16S rRNA gene sequence alone. Therefore, the additional marker is required to permit finer differentiation of closely related strains. One such marker readily differentiate the different strains of Phytoplasma are SecY gene, which encodes a protein translocase subunit. This represents one of the most promising markers for finer differentiation of Phytoplasma strains for delineating biologically and/ or ecologically distinct strains that often cannot be readily resolved by analysis of the 16S rRNA gene alone.37 The present study reports the identification and molecular characterization of Phytoplasma associated with sesame Phyllody from north India based on 16SrRNA and SecY gene sequence analysis.
Disease survey and sample collection
Roving survey was conducted during September to December, 2012-14 in Varanasi and Mirzapur districts of Uttar Pradesh, India to know the incidence and severity of Phyllody disease on sesame. During the survey, sesame plants exhibiting diverse symptoms were recorded. Incidence of Phyllody in sesame fields (% of plants with Phyllody symptoms) was estimated by visual inspection of around 1,000 plants in each field, following “W” pattern (crossing the rows). Disease incidence was calculated as the percentage of symptomatic plants to the total number of plants observed. The Phyllody disease samples were collected from the different farmers fields separating with a distance of 10kilomers, between them. A part of the samples was used for DNA isolation and the remaining sample was stored at -80oC for further use. The isolates collected from the different farmers fields were designated as SPP1, SPP2, SPP3 (Varanasi), SPP4, SPP5 and SPP6 (Mirzapur).
DNA extraction and PCR amplification of 16Sr RNA and SecY gene
Total nucleic acids were extracted from the leaf samples collected from both symptomatic and asymptomatic plants using cetyl-trimethyl ammonium bromide (CTAB) method.38 PCR amplification of 1.8kb 16S rRNA gene was carried out using Phytoplasma specific universal primer pair P1/P7.39,40 The Amplicons were re-amplified in the second round PCR reaction using more specific internal primers R16F2n/R16R2 as the procedure described for Nested-PCR with expected product size of 1.2kb.25,39 Further the SecY gene of Phytoplasma was amplified by SecYF2 and SecYR1.37 This proves to be useful for finer differentiation among diverse strains. Amplification was performed with 35 cycles of denaturation for 1 min at 94oC, primer annealing for 45s at 55oC and primer extension for 1mints 30s at 72oC, with initial denaturation at 94oC for 3 mins and final extension of 15min at 72 oC. The PCR reactions were carried out in a Gene Amp PCR system 9700 (PE Applied Biosystems, Foster City, CA) thermo cycler. PCR reactions were carried out in a volume of 25μL containing 100ng of DNA template, 0.5U Taq DNA polymerase (Fermentas, Germany), 2mM MgCl2 (Fermentas, Germany), 0.16mM dNTPs (Fermentas, Germany) and 0.3μM of each primer. PCR products were electrophoreses (1h at 80volts) in 0.8% agarose gel and stained with Ethidium bromide (10mg/mL) in Tris-borate-EDTA buffer (pH 8). Gels were visualized in a Gel documentation unit (Alpha InfoTech, USA). The Cyclic conditions and PCR reaction components were same for both direct and Nested-PCR, except the primers.
Cloning of PCR product and sequencing
The amplified products for primer pair P1/P7 (1.8kb size) and SecY gene (1.6kb size) were excised from the gel and purified by Gel extraction kit (Qiagen). The fragments were ligated into the pTZ57R/T vector (Fermentas, Germany) as the manufactures instructions. The vector was transformed into Escherichia coli DH5α competent cells (Invitrogen Disservices India Pvt. Ltd. at Bangalore).41 And recombinant clones were identified by restriction end nuclease digestion as well PCR amplification using primer pair R16F2n/R16R2 and SecYF2 and SecYR1 as described above. The selected clones were sequenced with automated sequencing ABI PRISM 3730 (Applied Biosystems) from Amnion Bioscience DNA Sequencing facility, Bangalore, Karnataka, India.
Restriction fragment length polymorphism analysis
The amplified nested-PCR product of 16S rRNA of six sesame phyllody isolates was digested with restriction enzymes such as AluI, EcoRI Taq1, HaeIII and HhaI42 which are used in finer classification of phytoplasma and their strains. Similarly the PCR amplified SecY gene product was digested with AluI, Taq1, RsaI and HhaI restriction enzymes as described by Lee37 for finer differentiation among diverse strains. The PCR-RFLP pattern of digested 16S rRNA and SecY gene was analyzed through electrophoresis with 2% agarose gel stained with Ethidium bromide (10mg/mL), using 0.5xTBE as running buffer. DNA bands were visualized in a UV transilluminator. PCR-RFLP patterns obtained were compared with previously described patterns.37,42
In-silico RFLP analysis
In-silico restriction analysis of R16F2n/R2 fragment of SPP isolate were performed using iPhyClassifier (http://www.ba.ars. usda.gov/data/ mppl/) software.38 The sequence was digested with 17 different restriction enzymes (AluI, BamHI, BfaI, BstUI (ThaI), DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI (MboI), MseI, RsaI, SspI and TaqI) and were plotted in a virtual 3.0% agarose gel. The Phytoplasma were routinely differentiated on the basis of 16S rRNA gene by means of RFLP analysis of nested PCR-amplified R16F2n/R2 fragment using a number of end nuclease restriction enzymes.42 As the RFLP pattern of each Phytoplasma is conserved. The virtual RFLP patterns with the key enzymes that distinguish from previously recognized group/subgroup patterns were made in iPhy Classifier. The virtual RFLP gel patterns of sesame Phytoplasma were compared with 16Sr II group Phytoplasma for finer differentiation from its existing members in the Ca. P. australasia.
Sequence analysis
To assess the taxonomic position of six sesame phyllody phytoplasma isolates, full length 16SrRNA and SecY gene sequence were queried using iPhyClassifier online tool.37,43 Further, sequences were subjected to BLAST, NCBI to search for similar sequences in the database. The related sequences obtained from the database were aligned using Crustal X method implemented in SEAVIEW program44,45 and used for the construction of phylogenetic tree through the neighbour joining method using MEGA 6.01 version software.46 With 1000 bootstrapped replications to estimate evolutionary distances between all pairs of sequences simultaneously. The nucleotide sequence identity matrixes for the sesame phyllody phytoplasma were generated using Bio edit Sequence Alignment Editor (version 5.0.9).46
Detection of recombination events
The phylogenic evidence for recombination was detected by aligning 16Sr RNA and SecY gene nucleotide sequences of different groups of phytoplasma retrieved from database and the sesame isolate (SPP1) using the neighbour-Net method, Splits-Tree version 4.3.47,48 This method depicts the conflicting phylogenetic signals caused by recombination as cycles within unrooted bifurcating trees. Recombination analysis was carried out using Recombination Detection Program (RDP), GENECOV, Boots can, Max Chi, Chimara, Si Scan and 3Seq integrated in RDP4 to detect the recombination break points.49 Default RDP settings with 0.05 P- value cut off throughout and standard Bonferroni correction were used.
Survey for the disease incidence
The survey was conducted two times during the crop growth period, one at flowering stage and another at pod development stage. The sesame Phyllody is very much prevalent in the districts of Varanasi and Mirzapur, Uttar Pradesh state of India. The disease incidence was ranged from 30-70 per cent in different farmer’s fields (Table 1). The Phyllody symptoms were observed in the field by visual inspection of around 1,000 plants, following “W” pattern (crossing the rows). During inspection the most common symptoms observed in flowering stage are yellowing, Phyllody (all floral parts into dark green leaf-like structures), floral proliferation, floral virescence, formation of dark exudates on foliage and floral parts. Whereas in case of pod development stage, plants are expressing symptoms of phyllody, seed capsule cracking, shoot apex fascination. The most common symptom observed across the fields are transformation of all floral parts into dark green leaf-like structures with vein clearing in different floral parts. Further, whole inflorescence become twisted, leaves are reduced in size and closely arranged on the top of the stem with very short intermodal length giving appearance of broom (Figure 1a). The places of survey, number fields surveyed, crop stage and disease symptoms observed on sesame plants in different farmer’s fields in Varanasi and Mirzapur districts are given the (Table 1).
No. |
Place |
No. of Filed Surveyed |
Stage |
Type of Symptoms |
Av.% Disease Incidence |
PCR |
Varanasi district |
||||||
1 |
Jayapur |
5 |
Flowering |
Phllody, floral proliferation, |
30-45 |
+ |
2 |
Jamuni |
4 |
,, |
Phllody, yellowing, |
20-30 |
+ |
3 |
Khaira |
2 |
,, |
Phllody, floral proliferation, dark |
35-38 |
+ |
4 |
Pachraho |
2 |
,, |
Phllody, floral proliferation, dark |
15-25 |
+ |
5 |
Marach |
6 |
,, |
Phllody, yellowing, |
35-40 |
+ |
6 |
Churavanpur |
4 |
,, |
Phllody, floral proliferation, dark |
35-40 |
+ |
7 |
Betapur |
2 |
,, |
Phllody, floral proliferation, dark |
26-35 |
+ |
8 |
Muradi |
2 |
,, |
Phllody, yellowing, |
15-20 |
+ |
9 |
Parsupur |
4 |
,, |
Phllody, yellowing, |
25-30 |
+ |
10 |
Tophapur |
2 |
,, |
Phllody, floral proliferation, dark |
35-45 |
+ |
11 |
Karsara |
5 |
,, |
Phllody, yellowing,floral proliferation, |
25-30 |
+ |
12 |
Patewa |
3 |
,, |
Phllody, yellowing, |
15-20 |
+ |
13 |
Hariharpur |
3 |
Pod |
Phllody, capsule cracking, |
30-35 |
+ |
14 |
Niyashipur |
4 |
,, |
Phllody, capsule cracking, |
35-40 |
+ |
15 |
Rajapur |
5 |
,, |
Phllody, capsule cracking, |
30-35 |
+ |
16 |
Tarapur |
6 |
,, |
Phllody, |
25-30 |
+ |
17 |
kachariya |
5 |
,, |
Phllody, |
30-35 |
+ |
18 |
Madhopur |
2 |
,, |
Phllody, capsule cracking, |
25-30 |
+ |
19 |
Badoni |
2 |
,, |
Phllody, |
25-30 |
+ |
20 |
Mathaldae |
8 |
Flowing |
Phllody, yellowing, |
15-20 |
+ |
21 |
Babatpur |
5 |
,, |
Phllody, yellowing, |
45-50 |
+ |
22 |
Mohansari |
4 |
,, |
Phllody, floral proliferation, dark |
35-45 |
+ |
23 |
Nakkupur |
2 |
,, |
Phllody, |
40-45 |
+ |
24 |
Kurhuan |
3 |
,, |
Phllody, |
25-30 |
+ |
25 |
Bachhaw |
3 |
,, |
Phllody, floral proliferation, dark |
15-20 |
+ |
26 |
Baharapur |
3 |
,, |
Phllody, yellowing, |
25-30 |
+ |
27 |
Kadichak |
5 |
,, |
Phllody, yellowing, |
35-45 |
+ |
28 |
Khagrajpur |
2 |
,, |
Phllody, |
25-30 |
+ |
29 |
Dhadorpur |
1 |
,, |
Phllody, floral proliferation, dark exudates |
15-20 |
+ |
30 |
Kanthipur |
2 |
,, |
Phllody, floral proliferation, dark |
30-35 |
+ |
31 |
Rajapur |
3 |
,, |
Phllody, yellowing, |
35-40 |
+ |
32 |
Belawan |
5 |
,, |
Phllody, floral proliferation, dark |
30-35 |
+ |
33 |
Sihorawan |
3 |
,, |
Phllody, |
25-30 |
+ |
34 |
Baburampua |
4 |
,, |
Phllody, |
30-35 |
+ |
35 |
Paniara |
2 |
,, |
Phllody, floral proliferation, dark |
25-30 |
+ |
36 |
Koelipur |
5 |
,, |
Phllody, yellowing, |
25-30 |
+ |
37 |
Gotawan |
2 |
,, |
Phllody, floral proliferation, dark |
15-Oct |
+ |
38 |
Akelwa |
3 |
,, |
Phllody, |
15-20 |
+ |
39 |
Aahim |
2 |
,, |
Phllody, floral proliferation, dark |
15-Oct |
+ |
40 |
Bandhawan |
2 |
,, |
Phllody, |
15-Oct |
+ |
41 |
Duhae |
1 |
,, |
Phllody, floral proliferation, dark |
20-25 |
+ |
Table 1 Survey for sesame phyllody in different location of Varanasi and Mirzapur in Uttar Pradesh
Detection of phytoplasma
All the six sesame phyllody samples collected from different farmers’ fields gave positive amplification in PCR for the universal primer pair P1/P7.40,50 Followed by nested PCR with R16F2n/R225,39 primers confirming the association of phytoplasma with them. No amplification was obtained from the non-symptomatic samples (Data not shown). The amplification with primer pair P1/P7 may result in no amplification or weak amplification. In order to rule out this error, the Nested- PCR was done to further confirm amplification (positive/negative) by direct PCR. Further, all the six samples gave amplification to primer pair SecYF2/SecYR1 designed to amplify the SecY gene of phytoplasma (approx. size 1.6 kbp). The amplified (for both primer pair P1/P7 and SecYF2/SecYR1) PCR products (1.8kb and 1.6kb) from six infected sesame samples were cloned and sequenced. The 16srRNA and SecY gene sequences of all the six isolates were found identical. Hence, one (SPP1) representative sequence of sesame isolates in the present study was deposited in the Gen Bank [Accession No: KF700083 (16srRNA), KT970076 (SecY gene)].
Sesame phyllody phytoplasma 16SrRNA and SecY gene sequence analysis
The isolate from Uttar Pradesh SPP1 sequence obtained in the current study was compared with 16SrRNA gene sequence of selected 62 known phytoplasma belonging to different groups and subgroups available the database. The sequence of SPP1 isolate shared nucleotide identity from 97.9 to 99.9% with sesame phyllody phytoplasma belongs to 16SrII peanut witches’-broom group (Table 1a). Within this group, it shared highest homology (99.5 to 99.9 %) with 16S rRNA sequence of sesame phyllody phytoplasma (KF322278, KF322275, KF322277, KF322279, KF429485, KF322273, KF322274 and AB690308) from Indian subcontinent, Chickpea phyllody-16SrII-D (FJ870549), Ca.P.australasia-16Sr II-D (Y10097) from Australia, Peanut witches-broom- 16Sr II-A(L33765) from Taiwan. Comparison of sesame phyllody within the subgroup of 16Sr II showed, nucleotide identity of 97.9 to 98.5% with Tomato witches-broom 16Sr II-D(HM584815), Picris echiodes phyllody- 16Sr 11-E (Y16393), Cactus witches-broom [EU099552 (16Sr II-J), EU099546 (16Sr II-L), EU099568 (16Sr II-G), EU099556 (16Sr II-F), EU099572 (16Sr II-K), EU099569(16Sr II-H), EU099551(16Sr II-I)], Crotalaria phyllody- 16Sr II-C (EF193355), Ca.P.aurantifolia- 16Sr II-B (U15442) of the 16Sr II Peanut WB group (Table 1b). The current classification criteria for phytoplasma based on16Sr RNA sequencing placed the Phytoplasma isolates as subgroups which share nucleotide identity of 94-100 percent and isolates as groups which share 80 and above per cent.25 The 16SrRNA gene sequence of Phytoplasma in the present study shares nucleotide identity of more than 94 per cent with members of peanut witches’-broom group (16Sr II), therefore it may be regarded as a member of peanut witches’-broom group (16Sr II). Similarly, the analysis of SecY gene showed that, the current isolate share nucleotide sequence identity between members of different Phytoplasma groups from 30.2 to 96.2% (Table 2 & 3). Further comparison of SecY gene of SPP1 isolate with members of different subgroups group of 16Sr II available in the database revealed highest nucleotide identity of 95.8 to 96.3% with sesame Phyllody (GU004362, AB703253) and Australian tomato big bud-16Sr II-D (GU004347) and lowest identity of 66.2 to 84.7% with Soybean Phyllody (GU004324), Picris echinoids (GU004348) Peanut witches-broom (GU004331) and Sesame Phyllody (GU004322). This indicates, the SecY gene isolated from sesame phyllody SPP1isolate belong to the subgroup 16Sr II-D and is more informative molecular tool for classification of closely related phytoplasma strains.
Phytoplasma Species |
Sub-Group |
Accession No. |
Country |
Sesame phyllody -Pali-Rajasthan |
16SrII |
KF429485 |
India |
Sesame phyllody - Kheda-2 Gujarat |
16SrII |
KF322274 |
India |
Sesame phyllody - Kheda-1 Gujarat |
16SrII |
KF322273 |
India |
Sesame phyllody -TKG-N32 |
16SrII |
KF322277 |
India |
Sesame phyllody – Meiktila |
16SrII |
AB690308 |
Myanmar |
Sesame phyllody -TKG-431 |
16SrII |
KF322278 |
India |
Sesame phyllody - TKG-421 |
16SrII |
KF322275 |
India |
Sesame phyllody - Kushinagar-2 UP |
16SrII |
KC920748 |
India |
Sesame phyllody - TKG-307 |
16SrII |
KF322279 |
India |
Sesame phyllody -TKG-JTS |
16SrII |
KF322276 |
India |
Tomato witches-broom |
16SrII-D |
HM584815 |
Saudi Arabia |
Ca. P. aurantifolia |
16SrII-B |
U15442 |
Oman |
Peanut witches-broom phytoplasma |
16SrII-A |
L33765 |
Taiwan |
Ca.P. austrlasia |
16SrII-D |
Y10097 |
Australia |
Crotalaria phyllody phytoplasma |
16SrII-C |
EF193355 |
Thailand |
Cactus witches'-broom phytoplasma |
16SrII-G |
EU099568 |
China |
Cactus witches'-broom phytoplasma |
16SrII-F |
EU099556 |
China |
Cactus witches-broom phytoplasma |
16SrII-H |
EU099569 |
China |
Cactus witches-broom phytoplasma |
16SrII-I |
EU099551 |
China |
Cactus witches-broom phytoplasma |
16SrII-J |
EU099552 |
China |
Cactus witches-broom phytoplasma |
16SrII-K |
EU099572 |
China |
Cactus witches-broom phytoplasma |
16SrII-L |
EU099546 |
China |
Picris echiodes phyllody phytoplasma |
16Sr11-E |
Y16393 |
Italy |
Ca.P.fraxini |
16SrVII |
AF092209 |
USA |
Ca.P.ulmi |
16SrV |
AY197655 |
USA |
Ca.P.palmae |
16SrVIII |
U18747 |
USA |
Ca.P.cynodontis |
16SrXIV |
AJ550984 |
Italy |
Ca.P.phoenicium |
16SrIX |
AF515636 |
Lebanon |
Ca.P.pruni |
16SrIII |
L04682 |
USA |
Ca.P.mali |
16SrX |
AJ542541 |
Italy |
Ca.P.asteris |
16SrI |
M30790 |
Michigan |
Ca.P.australiense |
16SrXIII |
L76865 |
Australia |
Pigeon pea witches’-broom |
16SrIX |
AF248957 |
USA |
Ash yellows |
16SrVIII |
AF189215 |
USA: New York |
Ca.P.braziliense |
16SrXV |
AF105315 |
USA |
Apple proliferation |
16SrX |
AF248958 |
Italy |
Chickpea phyllody |
16SrII |
FJ870549 |
Pakistan Faisalabad |
Clover phyllody |
16SrI |
AF222065 |
Canada |
Cactus witches'-broom |
16SrII |
AJ293216 |
China |
Clover yellow edge |
16SrIII |
AF189288 |
USA:Oregon |
Coconut lethal yellowing phytoplasma |
16SrIV |
AF498307 |
Jamaica |
Ca.P.trifolii |
16SrVI |
AY390261 |
Canada |
Fragaria multicipita phytoplasma |
16VI-G |
AF190225 |
Canada |
Ca.P.luffae |
16SrVIII |
AF353090 |
Taiwan |
Ca.P.oryzae |
16SrXI |
AB052873 |
Thailand |
Ca.P.solani |
16SrXII |
AJ964960 |
Spain |
Periwinkle virescence |
16SrXIII |
AF248960 |
Mexican |
Ca.P.brasiliense |
16SrXV |
AF147708 |
USA |
Ca.P. graminis |
16SrXV1 |
AY725228 |
Cuba |
Ca.P. caricae |
16SrXVII |
AY725234 |
Cuba |
Ca.P. americanum |
16SrXVIII |
DQ174122 |
USA |
Ca.P. castaneae |
16SrXIX |
AB054986 |
South Korea |
Ca.P. rhamni |
16SrXX |
X76431 |
Europe |
Ca. P.pini |
16SrXXI |
AJ632155 |
Spain |
Phytoplasma sp. strain |
16SrXXII |
Y14175 |
Nigeria |
Grapevine yellows |
16SrXXIII |
AY083605 |
Australia |
Sorghum bunchy shoot phytoplasma |
16SrXIV |
AF509322 |
Australia |
Tea witches broom |
16SrXXV |
AF521672 |
Australia |
Sugarcane phytoplasmaD3T1 |
16SrXXVI |
AJ539179 |
Mauritius |
Sugarcane phytoplasmaD3T2 |
16SrXXVII |
AJ539180 |
Mauritius |
Der bid phytoplasma |
(16SrXXVIII |
AY744945 |
Cuba |
Ca.P. malaysianum |
(16SrXXXII-A |
EU371934 |
Malaysia |
Table 1a 16srDNA Sequences of Phytoplasma employed in analysis
Phytoplasma Species |
Sub-group |
Accession No. |
Country |
Sesame phyllody phytoplasma |
16SrII |
GU004322 |
Thailand |
Sesame phyllody phytoplasma |
16SrII |
GU004362 |
Thailand |
Sesame phyllody phytoplasma |
16SrII |
AB703253 |
Myanmar |
Brinjal little leaf phytoplasma |
16SrVI-D |
GU004356 |
India |
Potato witches’-broom phytoplasma |
16SrVI-A |
GU004316 |
Canada |
Clover phyllody phytoplasma |
16SrVI-A |
GU004315 |
Canada |
Potato purple top phytoplasma -AK |
16SrVI-A |
GU004343 |
Alaska, USA |
Lucerne virescence phytoplasma |
16SrVI-A |
GU004318 |
France |
Vinca virescence phytoplasma |
16SrVI-A |
GU004317 |
California, USA |
Potato purple top phytoplasma -AK |
16SrVI-A |
GU004344 |
Alaska, USA |
Potato purple top phytoplasma -AK |
16SrVI-A |
GU004342 |
Alaska, USA |
Potato purple top phytoplasma -AK |
16SrVI-A |
GU004351 |
Alaska, USA |
Dry bean phyllody phytoplasma |
16SrVI-A |
GU004352 |
Washington, USA |
Dry bean phyllody phytoplasma |
16SrVI-A |
GU004353 |
Washington, USA |
Ash yellows phytoplasma |
16SrVI-A |
GU004329 |
New York, USA |
Milkweed yellows phytoplasma |
16SrIII-F |
GU004340 |
New York, USA |
Potato purple top phytoplasma-MT |
16SrIII-M |
GU004333 |
Montana, USA |
Clover yellow edge phytoplasma |
16SrIII-B |
GU004332 |
Lithuania |
Spirea stunt phytoplasma |
16SrIII-E |
GU004326 |
New York, USA |
Poinsettia branch-inducing phytoplasma |
16SrIII-H |
GU004328 |
USA |
Peach X-disease phytoplasma |
16SrIII-A |
GU004327 |
Canada |
Walnut witches-broom phytoplasma |
16SrIII-G |
GU004325 |
Georgia, USA |
Apple proliferation phytoplasma |
16SrX-A |
GU004335 |
Italy |
Mexican periwinkle virescence phytoplasma |
16SrXIII-A |
GU004336 |
Mexico |
Tomato big bud phytoplasma |
16SrI-A |
AY803178 |
Arkansas, USA |
Chrysanthemum yellows phytoplasma |
16SrI-A |
AY803170 |
Germany |
Hydrangea phyllody phytoplasma |
16SrI-A |
AY803181 |
Belgium |
Chrysanthemum yellows phytoplasma |
16SrI-B |
DQ787851 |
Italy |
Primrose virescence phytoplasma |
16SrI-B |
AY803176 |
Germany |
Clover phyllody phytoplasma |
16SrI-C |
AY803183 |
Germany |
Paulownia witches-broom phytoplasma |
16SrI-D |
AY803184 |
Taiwan |
Blueberry stunt phytoplasma |
16SrI-E |
AY803169 |
Michigan, USA |
Apricot chlorotic leaf roll phytoplasma |
16SrI-F |
AY803166 |
Spain |
Strawberry multiplier phytoplasma |
16SrI-K |
AY803180 |
Florida, USA |
Aster yellows phytoplasma |
16SrI-M |
AY803168 |
Germany |
Ipomoea witches-broom phytoplasma |
16SrI-N |
AY803182 |
Taiwan |
Peanut witches-broom phytoplasma |
16SrII-A |
GU004331 |
Taiwan |
Soybean phyllody phytoplasma |
16SrII-C |
GU004324 |
Thailand |
Picris echioides phytoplasma |
16SrII-E |
GU004348 |
Italy |
Australian tomato big bud phytoplasma |
16SrII-D |
GU004347 |
Australia |
Elm yellows phytoplasma |
16SrV-A |
AY197690 |
New York, USA |
Cherry lethal yellows phytoplasma |
16SrV-B |
AY197693 |
China |
Alder yellows phytoplasma |
16SrV-C |
AY197692 |
Germany |
Flavescence doree phytoplasma |
16SrV-D |
AY197685 |
Italy |
Rubus stunt phytoplasma |
16SrV-E |
AY197696 |
Italy |
American potato purple top wilt phytoplasma |
16SrXVIII-B |
GU004338 |
Nebraska, USA |
Stolbur-It phytoplasma |
16SrXII-A |
GU004355 |
Italy |
Pear declinev phytoplasma |
16SrX-C |
GU004363 |
Italy |
Coconut lethal yellows phytoplasma |
16SrIV-A |
GU004320 |
USA |
Candidatus Phytoplasma fraxini |
16SrVII-A |
GU004329 |
USA |
Table 1b SecY gene sequences of different phytoplasma employed in analysis
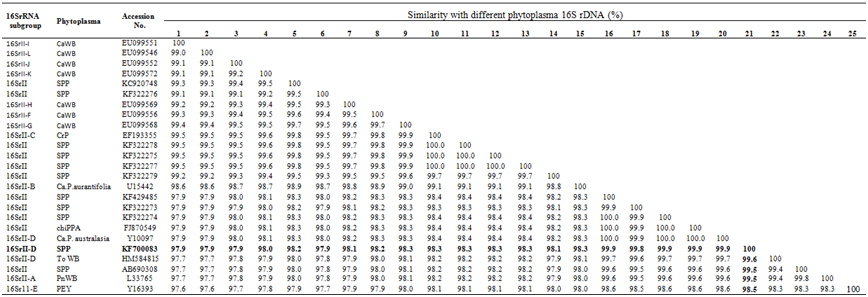
Table 2 Analysis of the sequence similarities among the 16SrRNA gene sequences from the Phytoplasma grouped in the group 16SrII available in database
The species are indicated as SPP: Sesame Phyllody Phytoplasma; Ca WB: Cactus Witches-Broom Phytoplasma; CrP: Crotalaria Phyllody; PnWB: Peanut Witches-Broom Phytoplasma; chiPPP: Chickpea Phyllody Phytoplasma; PEY: Picris Echiodes Phyllody Phytoplasma; To WB: Tomato witches-broom

Table 3 Analysis of the sequence similarities among the SecY gene sequences from the Phytoplasma grouped in the group 16SrII available in database
Phylogenetic analysis of 16SrRNA and SecY gene sequence analysis
The phylogenetic tree was generated by comparing the isolate SPP1 16SrRNA gene sequence characterized in the present study with other selected 62 phytoplasmas belongs to different groups and subgroups infecting different hosts sequences, which are available in the Gen Bank database (Figure 1). The pairwise similarity analyses showed that the newly characterised isolate SPP1 is grouped with previously identified sesame phyllody (KF429485, KF322273, KF322274 and AB690308), Tomato witches-broom 16Sr II-D (HM584815), Chickpea phyllody-16Sr II-D (FJ870549), Peanut witches-broom- 16Sr II-A (L33765) and Ca.P.austrlasia-16Sr II-D (Y10097) belonged to the members of peanut witches’-broom group (16SrII) infecting different crops in Indian subcontinents, Australia and Saudi Arabia (Figure 2a). The analysis showed Indian sesame infecting phytoplasma form a monophyletic cluster with Asian-Australasian- Saudi Arabia origin phytoplasma and established the close relationship between 16SrII-A and 16SrII-D. The analysis also showed that the oligo nucleotide sequences complementary to unique regions of the 16SrRNA 5’-TAAAAGGCATCTTTTATC- 3’ and 5’-CAAGGAAGAAAAGCAAATGGCG AACCATTTGTTT-3’ of isolate SPP1 phytoplasma was similar to the16SrII peanut witches’-broom group. The similarly, phylogenetic tree was generated by comparing the isolate SPP1 SecY gene with other 51 phytoplasma infecting different host are belongs to different groups and subgroups (Figure 1). The results revealed that, the SecY gene of isolate SPP1 is more closely clustered with sesame phyllody (GU004362, AB703253), Australian tomato big bud-16Sr II-D (GU004347) and Peanut witches-broom16Sr II-A (GU004331) belongs to group of 16Sr II (Figure 2b). The analysis showed Indian sesame infecting phytoplasma form a monophyletic cluster with Asian-Australasian origin phytoplasma and established the close relationship between 16Sr II-A and 16Sr II-D.

Figure 2a Phylogenetic trees based on sequences of 16SrRNA (a) and SecY gene (b) from sesame phyllody Phytoplasma isolate SPP1 with other Phytoplasma strains using Neighbor-joining algorithm. Horizontal distances are proportional to sequence distances, vertical distances are arbitrary. The trees are unrooted. A bootstrap analysis with 1000 replicates was performed and the bootstrap percent values more than 50 are numbered along branches.
In-silico RFLP analysis
Analysis of the isolate SPP1 sequence with online tool iPhyClassifier indicated that the virtual RFLP pattern derived from the query of F2nR2 fragment of 16S rDNA sequence was identical (similarity coefficient 1.00) to the reference pattern of 16Sr group II and subgroup D (Gen Bank accession: Y10097, Ca. P. australasia-16SrII-D). The analysis further confirmed that Phytoplasma isolate SPP1 from sesame is belongs to 16Sr group II and subgroup16SrII-D.
RFLP analysis of 16SrRNA and SecY gene
The PCR amplified F2nR2 and SecY gene fragments of sesame phyllody isolates (SPP1, SPP2, SPP3, SPP4, SPP5 and SPP6) were digested with restriction endo nucleases, which are used in classification of phytoplasmas.37,42,50 The restriction patterns of samples collected from different farmers fields were similar indicating the phytoplasma associated with sesame in different places of Varanasi and Mirzapur were identical and belongs to the peanut witches’-broom group (16Sr II) (Figure 3a & 3b).

Figure 3a 16sRNA, Restriction fragment length polymorphism (PCR-RFLP) patterns of 16S rRNA (a) and SecY gene (b) from Indian sesame Phytoplasma isolates amplified by PCR using primers R16F2/R16R2 and SecYF2/SecYR1. DNA products were digested with restriction end nucleases AluI, EcoRI Taq1, HaeIII and HhaI for 16S rDNA and AluI, Taq1, RsaI and HhaI for SecY gene. Lane M: Molecular marker 1kb ladder; lane1, SPP1; lane 2, SPP2; lane 3, SPP3; lane 4, SPP4; lane 5, SPP5; lane 6, SPP6.
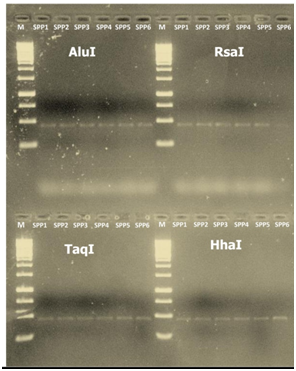
Figure 3b Neighbor-Net generated for the 16SrRNA (a) and SecY gene (b) of phytoplasma isolate SPP1 from sesame plant with other Phytoplasma strains has shown significant signals for phylogenic conflict indicatingas recombinant Phytoplasma.
Neighbor-net and recombination analysis of 16S rRNA and SecY gene of sesame phyllody
The neighbor-net analysis was carried out by aligned sequences of 16S rRNA and SecY gene of diverse groups phytoplasmas31,37 along with the 16S rRNA and SecY gene of isolate SPP1 using split tree program. The results revealed the extensive network of evolution in 16Sr II group/subgroups and SecY gene with other groups of phytoplasma indicating recombination in 16S rRNA and SecY gene of sesame phyllody phytoplasma, The split decomposition analysis showed a “rectangular” network structure suggesting sesame phytoplasma belong to 16SrII group/subgroupsand distinct from all other groups of phytoplasma. Bifurcation between sesame Phytoplasma belong 16Sr II group/subgroupsand other groups of phytoplasma in the split graph were similar to that of phylogenetic analysis. A comprehensive analysis of recombination using RDP3 based on the alignment of sequences of 16Sr II group/subgroups of Phytoplasma and other groups of Phytoplasma available in the database was carried out. The analyses revealed the evidence for inter species recombination in isolate SPP1 infecting sesame reported here with most of the part of the 16SrRNA F2nR2 fragment 414- 1643nt (P-value=9.518X10-26) was descended from Ash yellows-[16Sr VIII] (AF189215) and Apple proliferation-[16Sr X] (AF248958). In case of SecY gene, most of the part 1663- 23nt (P-value=2.748X10-17) was descended from Peanut witches'-broom- 16Sr II-A (GU004331) and aster yellows 16Sr I-A (GU004345) to emerge as a new strain of sesame phytoplasma.
Phytoplasma diseases are major production constraints of economically important field crops, oilseed, vegetables, fruit crops, ornamental plants, timber and shade trees.16 Their incidence is increasing day by day with novel symptomatology, uncertain etiology and diseases with diverse geographic distribution in the recent years.51 Incidence of the phtytoplasma diseases reported from the different parts of world suggests their ubiquitous presence.12,13,20,52,53 In India, the symptomatology of sesame phyllody dates back to several decades.54 However, identification of the exact species associated with that was lacking. Recently, it was identified as Ca. P. asteris (16Sr I group) by Klein.55 The current study revealed the presence of sesame Phytoplasma in the north-eastern parts of Uttar Pradesh, India with a considerable amount of incidence (35-50%) resulting in economic loss of the crop. Different detection tools based on nucleic acid such as PCR and N-PCR were available for the detection of Phytoplasma worldwide.13,55 And were employed in the management of the diseases. There was no difference between the incidences of Phytoplasma recorded based the symptoms observed in the field and molecular detection collected samples for by PCR. Phyllody causing Phytoplasma in several crops evolved independently and resulted in different groups. Worldwide, Phyllody disease in sesame was reported to be caused by three distinct phytoplasma groups viz. aster yellows, peanut witches’ broom and clover proliferation group.19,31,56 Some of the species most prevalent are Ca. P. asteris (16Sr I -B) from Myanmar.56 Peanut witches’ broom subgroup (16Sr II-D) from Pakistan and Oman.30,57 Peanut witches’ broom subgroup (16Sr II-A) from Thailand,19 Ca. P. trifolii’ subgroup (16Sr VI-A) from Turkey.19 In the present investigation, we have identified and classified Phytoplasma infecting sesame in north- eastern based on16SrRNA gene sequence and In-silico restriction analysis using iPhyClassifier online tools.43 The evidence suggests that, Phytoplasma SPP1 isolate causing sesame phyllody in north- eastern India is a member of 16Sr II-D subgroup belongs to the Peanut witches’ broom group. Further, the strain of phytoplasma associated with sesame in Varanasi and Mirzapur was identified by digestion of F2n/R2 fragment using five restriction enzymes42 And four restriction enzymes for SecY gene37 Those are used in the classification of Phytoplasma into groups and subgroups. The restriction pattern of Phytoplasma samples collected from different fields was identical, which indicates that, the same Phytoplasma is responsible for causing Phyllody in different locations. The RFLP patterns of every Phytoplasma is conserved, unknown Phytoplasma were identified by comparing the patterns of the unknown with the available RFLP patterns for known Phytoplasma without co-analyses of all reference representative Phytoplasma.42,58 It provides a reliable means for the differentiation of broad array of Phytoplasma and has become the most comprehensive and widely accepted Phytoplasma classification system.51 Recombination plays a significant role in creating genetic diversity within prokaryotic and eukaryotic virus populations.46,59
The most of the part of the 16SrRNA F2nR2 fragment of sesame Phytoplasma isolate SPP1 infecting sesame was known to be descended through inter species recombination with Ash yellows-[16Sr VIII] (AF189215) and Apple proliferation-[16Sr X] (AF248958) in 16sRNA. Whereas in case of SecY gene, most of the part is descended from Peanut witches'-broom- 16Sr II-A (GU004331) and aster yellows 16Sr I-A (GU004345) to emerge as a new strain of sesame Phytoplasma. Similarly, EC-DNA isolated from wild-type line (OY-W) and mild-symptom line (OY-M) of onion yellows Phytoplasma has encoded a geminivirus like Rep and a putative single-stranded-DNA-binding protein (SSB). The EC-DNA of wild-type line (OY-W) and mild-symptom line (OY-M) have intermolecular recombination between EC-DNAs in Phytoplasma.60 Recombination in extra-chromosomal DNA (EC-DNA) plays a major role in creating genetic diversity in Phytoplasma and provides the potential for rapid adaptation to new environmental conditions. This report added one more member of 16Sr IID subgroup from Peanut witches’ broom group in addition to, two Phytoplasma strains belonging Ca. P. asteris (16Sr I group) are responsible for causing sesame Phyllody in India. Further, the member of this Phytoplasma subgroup infecting chickpea have been identified in India and Pakistan.57,61–63 This clearly revealing the rapid expansion of host range by Phytoplasma belonged to 16SrII subgroup.
The authors are grateful to the Director of Indian Institute of Horticultural Research, Bangalore and Indian Institute of Vegetable Research, Varanasi, for providing research facilities and his keen interest in this study.
The authors declare that they have no conflict of interest.

©2017 :, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.