Advances in
eISSN: 2373-6402


One of the most represented group of Pathogenesis Related (PR) genes are those of the PR-10 class. PR-10 proteins are members of multi genic family, and they often occur in clusters at specific loci following gene duplication and amplification events. To date, large number of PR-10 genes have been cloned and characterized in different species in response to abiotic and biotic stress. This review is focused on recent studies that have described the role, distribution and structure of PR-10 genes in plant genomes. Recent findings have provided insights into the functional roles of PR-10 proteins as ribonuclease, as cytokinin-specific binding proteins, a mammalian lipid transport and plant abscisic acid (ABA) receptor proteins, or as enzyme, (S)-norco claurine synthase. PR-10 proteins are differentially expressed in the presence of different signaling molecules, biotic stresses such as fungal, viral and bacterial pathogens and a number of abiotic stresses. The possibility to use this knowledge for genetic improvement of plant resistance to pathogens through classical breeding approach or transgenic technology is discussed.
Keywords: pathogenesis related proteins, ribonucleases, multigene, rnase, abiotic, biotic, agrobacterium
ABA, abscisic acid; LRR, lucine rich repeat; NCS, (S)-norcoclaurine synthase; ORF, open reading frame; PR10, pathogenesis related class 10
Plant growth and survival is always influenced by several factors including abiotic and biotic stresses. Plants respond to these factors by inducing their defense mechanism which includes expression of several effectors, receptors, signaling and protective molecules. One of the most commonly induced proteins during plant defense mechanism is pathogenesis related (PR) protein. Accumulation of PR proteins is an integral component of innate immune responses in plants during pathogen attack or under abiotic stress conditions. The PR proteins not only accumulate locally in the infected leaf, but are also associated with the development of hypersensitive response (HR) or systemic acquired resistance (SAR) against infection by fungi, bacteria and viruses.1,2 The PR proteins are grouped into 17 families depending upon their primary structure, serological relationships and biological activities.3 Different families of PR proteins exhibit different antimicrobial and secondary metabolic enzyme activities, for example chitinases (PR3, PR4, PR8 and PR11),4,5 β-1, 3-glucanase (PR2),4 osmotin with thaumatin-like protein (PR5), RNase (PR-10), defensins (PR12),6 thionin (PR13), lipid-transfer protein (PR14) and oxalate oxidase (PR15 and 16).7-11 Most of the PR protein families are extracellular in nature, but some of the PRs are found in the cytoplasm also, abundantly in the vacuole.3 The role of different types of PR proteins during abiotic and biotic stresses and their defense responses in plants are very well documented in literature; however, their mechanism of action is sparsely described. The PR-10 family is the largest among all different classes of PR10 proteins, with more than 100 members reported across more than 70 plant species.3 This review article will summarize the current status, structural and functional diversity of PR-10 proteins with special emphasis on their role in abiotic and biotic stress tolerance.
The PR-10 class of PR proteins was first described in parsley and referred as ‘classic’ PR-10 proteins.12 PR-10 proteins are ubiquitous proteins that have been identified in a number of dicot and monocot plant species. They are small, slightly acidic and resistant to proteases. PR-10 proteins are classified as intracellular PR (IPR) proteins and are present in cytoplasm because they lack signal peptide. They are closely related to a group of major tree pollen allergens and food allergens based on sequence homology to classic PR-10 proteins (~50% identity). The common allergens found in birch pollen,13 celery,14 apple,15 peanut16 and tomato17 are included in the PR-10 class. Most PR-10 genes share an open reading frame (ORF) from 456 to 489bp (154-163 amino acids) which is interrupted by an intron of 76-359bp at a highly conserved position.3
Amino acid sequence alignments of PR-10 proteins clearly show the most divergent and most conserved segments (Figure 1). This ORF codes for a small protein with conserved sequence features such as a Glycine-rich loop or GXGGXGXXK motif (aa 47-55), a signature motif of PR-10 proteins which is conserved even in distant homologs. This motif has remarkable sequence similarity to P-loop, the Bet v 1 motif (IPR000916) characteristic of proteins from the Bet v 1 super family and three amino acids E96, E148 and Y150 (as positioned in Bet v 1) are possibly involved in ribonucleasic activity.18 P-loop, is a phosphate-binding loop found in nucleotide binding proteins.18 However, PR-10 proteins do not have affinity for ATP and the glycine-rich loop is conformationally different from the P-loop.19,20
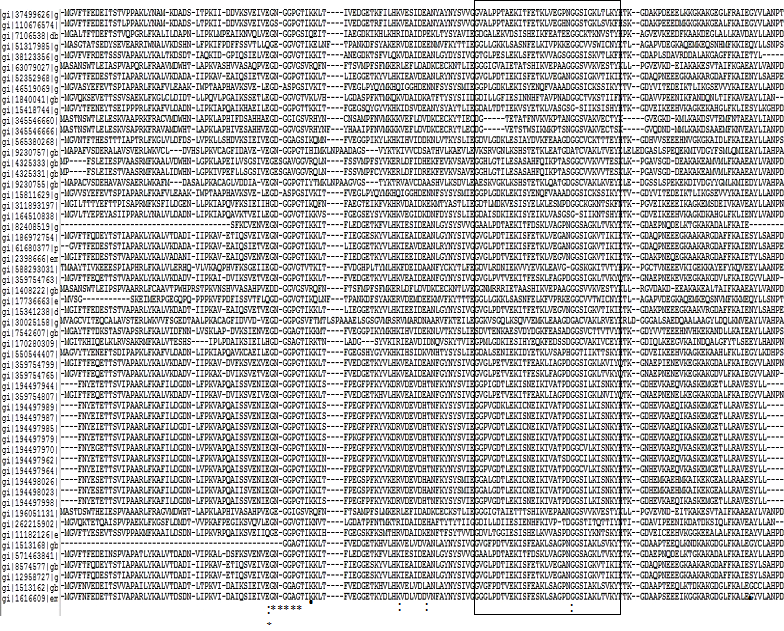
Interestingly, the glycine-rich loop is the most rigid structural element in the PR-10 fold despite being glycine rich. A characteristic START-like domain (IPR023393), an alpha/beta sandwich structural domain is found in a wide variety of PR protein families. Bet v1 and PYR/PYL/RCAR domains typically bind phytohormones such as brassino steroids, cytokinins and abscisic acid. Superposition of the PR-10 structures reveals very significant structural differences, mainly at the C-terminal helix a3, displaying different axial shifts as well as a variable degree of deformation at the center and at its N-terminal connection with loop L9.21 The internal cavity formed with the participation of a3, displays a remarkable variability in terms of the volume.
PR-10 genes are multi gene families having low intra specific variation but higher inter specific variation which makes them interesting phylogenetic markers.22 For example, at least five PR-10 genes in pea,23 eighteen Mal d 1 genes in apple,24 ten Bet v 1 genes in birch,25 eight Fra a 1 genes in strawberry,26 six PR-10 genes in Solanum surattense,27 eight in yellow lupine,28 five in rice,29 and eight Pru p 1 and Pru d 1 genes in peach and almond, respectively,30 have been identified.
They also tend to form physical clusters on specific chromosomes, e.g. in apple,24 peach30 and poplar31 PR-10 genes. Lebel et al.,22 found that thirteen out of the seventeen Vitis vinifera PR-10 sequences are present on the chromosome in direct orientation suggesting that most copies were produced by unequal crossing over events, as described in Arabidopsis and rice,.32 Gene duplication events in the genome evolution process make new copies of a gene which may undergo modifications resulting in functional diversification.33 These kinds of events are significant source of evolution in plants, however, most of the times gene copies produced by duplication are rapidly lost through pseudo genisation. Therefore, only a part of numerous homologous sequences coexisting in a genome are functional genes.
Another important aspect of PR-10 evolution is evident from differential patterns of expression among the different plant organs, i.e. root, leaf, stem and peduncle, indicating that the transcripts may represent functionally divergent genes.34,35 Furthermore, some PR-10 proteins are constitutively expressed in plants while some are induced only under biotic stress, abiotic stress or during plant development, emphasizing functional diversification.19,28 The silencing of MtPR-10-1 from Medicago truncatula led to the induction of a new set of PR proteins after infection with Aphanomyces euteiches,36 suggesting that there is a relationship between PR-10 and other PR proteins.
Structural and functional diversity: decoy strategies to fine tune the defense
PR-10 proteins are involved in many aspects of plant development, growth and defense but their molecular function is still unclear. Various roles for PR-10 proteins have been inferred, such as involvement in enzymatic processes, secondary metabolite biosynthesis, antimicrobial processes, storage, membrane binding, transport, phyto hormone and other hydrophobic ligand binding. However, most of the studies exploring PR-10 functions were conducted in vitro.37–43
A protein with ribonuclease activity was isolated from callus cell culture of Panax ginseng showing ~60–70% sequence identity with two intracellular PR proteins from parsley, but did not show any homology with other known ribo nucleases.37 The RNase activity of PR-10 proteins was also detected in Bet v 1 and BpPR-10c from birch.38,39 LaPR-10 from white lupin,40 LlPR-10.1B from yellow lupine,20 BpPR-10c from birch,41 GaPR-10 from cotton,42 SPE16 from Pachyrrhizus erosus,43 CaPR-10 from hot pepper,44 SsPR-10 from Solanum surattense,27 AhPR-10 from peanut,45 and PsPR-10.1 and PsPR-10.4 from pea.46–48 PR-10 proteins exhibiting RNase activity inhibit the growth of pathogen through direct cytotoxic impact on pathogen cells, possibly participating in the induction of plant cell apoptosis and development of hypersensitive reactions.49
Despite a number of studies associating RNase and antimicrobial activities of PR-10 proteins in the plant immune responses, tissue-specific expression of PR-10 gene during plant growth and development needs critical evaluation to determine the role for PR-10 proteins. While the selective RNA degradation activity may be critical to controlling the transcriptional burst in response to molecular events leading to stress perception or a downstream hypersensitive/apoptotic response essential to the containment of infection foci, it may also be directly responsible for arbitration of an invading pathogen.
PR-10 proteins behave as ribonucleotide binding proteins (RBP) and take part in virus resistance via binding to viral RNAs.50 Structural analysis of PR-10 indicated that it has quite diverse sequences as well as highly conserved sequences. PR-10 family has highly conserved regions including a specific domain (KAXEXYL), and the glycine-rich motif (GXGGXGXXK), which is known as a RNA binding site, but whether these sites have specific binding affinity to target RNA is not clear as PR-10 is also known to be involved in defense functions during a variety of abiotic and biotic stresses.21,30,46 PR-10 proteins have been reported to have several functions but there is no general function common to all members of this class. It is likely that the post translational modifications such as phosphorylation of the protein provide specificity for target RNAs, which in turn delimit potentially dangerous unspecific RNase activity.44
One of the member of PR-10 family, CaPR-10 isolated from hot pepper (Capsicum annuum), showed phosphorylation.44 Phosphorylation lead to enhanced ribonucleolytic activity against viral RNAs upon Tobacco Mosaic Virus (TMV) infection showing its direct involvement in plant defense.44 Some PR-10 proteins from Arachis hypogaea were shown to be phosphorylated but their role in RNase activity was not shown.51 A report shows that phosphorylation of CaPR-10 is enhanced by leucine-rich repeat 1 (LRR1) protein.10 However, Pungartnik et al.,52 demonstrated no effect of phosphorylation on the RNase activity or substrate specificity in the cocoa TcPR-10 protein.
The PR-10 protein from Theobroma cacao, TcPR-10 showed both antifungal activity against Moniliophthora perniciosa, and in vivo ribonuclease activity.52 Although non-specific effects of the PR-10 family were observed, the possibility of helper proteins for specific binding of target RNAs, such as viral or host RNAs of PR-10 proteins, cannot be overruled.21
Recently, Choi et al.,10 investigated a cytosolic interaction of CaPR-10and LRR1, an innate immune receptor recruited in response to pathogen attack. Compromised cell death mediated-defense signaling as observed in transgenic pepper infected with avirulent Xanthomonascam pestris pv. Vesicatoria after suppression of cytosolic PR-10/LRR1 interaction.
On the contrary, enhanced resistance to P. syringae pv. Tomato and Hyaloperonospora arabidopsidis was noticed under heterologous overexpression of PR-10/LRR1 in transgenic Arabidopsis, thus, corroborating the role for PR-10 proteins in conjunction with LRR1 during HR.10 However, the mechanism of CaPR-10-LRR1 interaction-mediated defense and how CaPR-10 recognizes the host RNAs are still unclear. On a similar note, the interaction between another family of PR proteins (PR4b) and LRR1 was demonstrated in hypersensitive cell death and defense response in pepper by Hwang et al.53 To investigate the role of three conserved residues Glu96, Glu148 and Tyr150 (ginseng ribonuclease sequence) in the RNase activity, site-directed mutagenesis of those residues was performed including some positions within the glycine-rich loop. The RNase activities of SPE16 and GaPR-10 are affected to a greater extent when residues of the C-terminal helix are substituted, while in the case of AhPR-10 major effects are seen with mutagenesis at the glycine-rich loop. An elevated level of PsPR-10.4 activity is observed when Glu148 is mutated to alanine and a decreased level is observed with an H69L mutation.48
Site directed mutagenesis of the peanut AhPR-10 protein deteriorated the RNAse and antifungal activities without any discernible effect on protein internalization by fungal mycelium of Fusarium oxysporum and Rhizoctonia solani in a hyphal extension inhibition assay.45 However, Biesiadka et al.,20 reported that despite having a high level (76.8%) of identity and sequence conservation at the RNase-relevant positions in two yellow lupine LlPR-10.1A and LlPR-10.1B proteins, only LlPR-10.1B showed RNase activity. Therefore, it is presumed that RNase activity is found in some PR10 proteins, but this is not a general property of this class of PR proteins.
Cytokinins, a class of plant growth phytohormones, have also been accepted as integral components of plant defense repertoire and abiotic stress responses.54 A subclass of PR10 proteins has been structurally confirmed as cytokinin-specific binding proteins (CSBPs) despite having marginal (<20%) sequence identity.55 Some of the classic PR-10 proteins were found to form complexes with brassinosteroid analogs,56 flavonoids57 and cytokinins.58 Constitutive expression of a ribonuclease-active pea PR-10 protein (PR-10.1) gene in Brassica napus seedlings enhanced endogenous cytokinin pool while promoting seedling germination and growth rates under saline conditions.46 Krishnaswamy et al.,59 suggested that PR-10 proteins may modulate cytokinin levels through an uncharacterized mechanism, which may include the degradation of tRNAs containing cytokinin moieties. Interestingly, an evolutionary ancient and versatile polyketide cyclase/dehydrase-like signature domain (polyketide_cyc, Pfam: PF03364) is found in PR-10 proteins, which may be involved in the binding of cytokinins, flavonoids and steroids across cellular aqueous environments.21 Zubini et al.,60 have investigated the possible role of the two Pru p 1 isoforms in the defense response of peach to the fungal pathogen Monilinia spp. The RNase activity is different for the two proteins, and only that of Pru p 1.01 is affected in the presence of the cytokinin zeatin, suggesting a physiological correlation between Pru p 1.01 ligand binding and enzymatic activity. The difference in binding activity pointed towards the differences in the binding pockets based on homology modeling.
PR-10 proteins have structural and sequence homology with mammalian lipid transport and plant abscisic acid receptor proteins and are predicted to have cavities for ligand binding.61 A large internal Y-shaped hydrophobic cavity, as determined by three-dimensional structure of PR-10 proteins could be liable for transport of a polar ligands such as fatty acids, flavonoids, cytokinins or brassino steroids in the intracellular spaces.62 The diverse roles predicted for PR-10 proteins in the plant immune system should have consideration of discernable modifications of the structure and shape of this cavity allowing to bind different ligands.20,63
In a recently study, three new members of the PR-10 family, the Fra a proteins, have been identified in strawberry in response to the flavonoid biosynthesis pathway, which is essential for the development of color and flavor in fruits and it was suggested that Fra a proteins could act as transporters or “chemical chaperones” binding to flavonoid intermediates so that they are available to processing enzymes.61 Furthermore, structural comparisons of the apo forms of Fra a 1E and the Fra a 3-catechin complex indicates that Fra a proteins show significant flexibility in the loop regions surrounding the cavity (loops L3, L5, and L7) and ligand-binding induces important conformational changes suggesting an important role of PR-10 proteins in control of secondary metabolic pathways.
The discovery of a PR-10 homolog with unique organ/tissue-specific expression in the tapetal cells during anther development suggests a potential role in the sporo pollen in pathway for these proteins.64 An enzyme (S)-norcoclaurine synthase (NCS) which is involved in benzyl iso-quinoline alkaloid biosynthesis, catalyzing a Pictet–Spengler condensation of dopamine and 4-hydroxy phenyl acetaldehyde to (S)-norcoclaurine share 28%–38% sequence identity with classic PR-10 proteins.65,66 Four NCS enzymes namely Tf NCS from Thalictrum flavum, Ps NCS1 and Ps NCS2 from Papaver somniferum, and Cj PR10A from Coptis japonica share substantial identity with PR10 and Bet v1 proteins.66 Similarly, the phenolic oxidative coupling protein (Hyp-1) from Hypericum perforatum which catalyze the condensation of two emodine molecules to the bioactive naphtha dianthrone hypericin, shows approximately 40% sequence identity with classic PR-10 proteins.67–69
Signaling nodes: PR-10 in response to signaling pathways
Phytohormones such as abscisic acid (ABA), ethylene (ET), jasmonic acid (JA) and salicylic acid (SA) are major signaling molecules in plants during the stress response, and their involvements during induction of PR10 proteins has been investigated in various studies.70 In general, SA is an important signal for general defense responses and especially for attack by bio trophic pathogens in so-called systemic acquired resistance (SAR) and the JA/ET signaling pathway is involved in responses to wounding and abiotic stresses such as drought and high salinity and also in the defense signaling against necrotrophic pathogens.71,72 ABA has a crucial role in responses to plant growth and development as well as in wide range of abiotic stresses, including drought, salt and cold. A diagrammatic representation of the expression of defense related proteins and transcription factors in response to signaling molecules are shown in Figure 2.
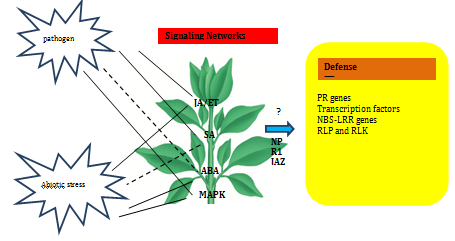
Expression of a rice PR10 protein, RSOsPR10 is regulated antagonistically by JA/ET and SA signaling pathways in response to environmental stresses.72 Accumulations of JIOPR1073 and OsPR1074 transcripts were observed on application of JA and SA in rice leaves. The folding canon of PR-10 proteins is found in the ABA receptor family known as PYR/PYL/RCAR (pyrabactin resistance/PYR-like/regulatory component of ABA response).21 Over-expression of a rice transcription factor, OsWRKY30, activates the expression of LOX, AOS2, PR3 and PR10 genes, increases endogenous JA levels and confers resistance to the rice fungal pathogens Rhizoctonia solani and Magnaporthe grisea.75 Following ethylene treatment enhanced levels of accumulation of PR10 transcripts were observed in OsPR10a from rice76 and Pg1 from ginseng.77
Two alfalfa PR10 genes, MsPR10.1A and MsPR10.1B, were responsive to ethylene and ABA.78 Analysis of the root proteome of moderate susceptible Medicago truncatula in response to infection by the oomycete root pathogen Aphanomyces euteiches and the abundance levels of one group of ABA-responsive proteins (ABR17) of the PR-10 class were observed indicating that ABA-mediated signaling is involved in PR protein induction for disease resistance.79 Therefore, despite the fact that the mechanism of interaction between signaling molecules and PR-10 proteins remains largely unknown, the results of a number of studies suggest that PR10 expression is triggered by the application of signaling molecules and that this response is important in host resistance.
Abiotic and biotic stresses: PR-10 response
Plants are responsive to environmental factor and may adapt to certain amount of abiotic and biotic stresses by activating their survival strategies through changes in biochemical and physiological pathways. Activation of the plant immune system that allows survival of plants in response to these extreme stress regimes is important. The PR-10 genes are one of the important components of the plant growth and developmental system and are differentially regulated by various environmental stimuli such as pathogen attack and/or abiotic stresses. Some PR-10 proteins are shown to possess antifungal activity such as, AhPR-10 of Arachis hypogaea45 and TcPR-10 of Theobroma cacao52 through RNase activity and internalization of fungal mycelium. Other PR-10 proteins that possess antifungal activity are SsPR-10 from Solanum surattense.,27 maize PR-10 proteins,9 CsPR-10 from Crocus sativus80 and JcPR-10a from Jatropha curca.81
A study by Soh et al.,11 also demonstrated enhanced expression and longevity of PR-10 gene transcripts in a disease-resistant pepper cultivar in response to the fungal pathogen Colletotrichum acutatum. In a recent study by Fan et al.,82 a novel PR-10 Protein Gly m 4l, was found to increases resistance upon Phytophthora sojae infection in soybean (Glycine max [L.] Merr). Gly m 4l transcripts were increased by SA stress, but relatively low under MeJA and ET treatments, and almost decreased with ABA and GA3 treatments, therefore it was speculated that Gly m 4l might play a key role in soybean plants resistance to P. sojae mainly depending on SA signaling.
Some PR-10 proteins also show antibacterial and antiviral activity. Ocatin inhibits the growth of phytopathogenic bacteria, such as Agrobacterium tumefaciens, Agrobacterium radiobacter, Serratiamarcescens and Pseudomonas aureofaciens.83 The PR-10 proteins from maize, ZmPR-10 and ZmPR-10.1 have antibacterial activity against bacteria P. syringae.9 Antiviral activity of pepper CaPR-10 was shown to degrade viral RNA of tobacco mosaic virus.44
Antinematode activity has been reported for PR-10 proteins. The CpPRI from Crotalaria pallid roots shows nemato static and nematicide effects against root-knot nematode Meloidogyne incognita by inhibiting the papa in-like enzymes present in the digestive tube and the cuticles of the pathogens.84 In addition to papa in inhibition, CpPRI was observed to internalize and diffuse over the entire body of juvenile M. incognita nematodes in fluorescence based assay.84 In another study, transcripts of genes encoding PR-10 (SAM22) were increased 5- to 10-fold after 12days of infection and remained high even 10weeks after infection.85 Similarly, PR-10 expression was higher in resistant pine trees than in susceptible pine trees at 7 and 14days post inoculation with the pine wood nematode (PWN) Bursaphelenchus xylophilus.86 Synchronized expression of PR-10 with peroxidase in resistant trees indicates this gene may be induced by reactive oxygen species (ROS) such as H2O2 or it may act as a proteinase against some enzymes such as cellulases, beta-1,3-glucanase, and pectate lyases which are secreted from PWN.87
PR-10 proteins have been shown to be transcriptionally responsive across a large range of abiotic stress environments such as drought, salinity, low and high temperatures, heavy metals, wounding and UV exposure,9,72,88 Several proteins with similarities to the PR-10 family members were identified through two dimensional gel electrophoresis, which were up-regulated in peanut callus cultures subjected to salt stress.51 Transgenic overexpression of one peanut salinity-induced PR-10 gene (AhSIPR10) in tobacco exhibited enhanced tolerance to salt, heavy metal (ZnCl2) and mannitol-induced drought stress.88 The expression of CcPR-10 transcripts was induced by wounding and jasmonic acid treatments as well as by armyworm (Spodoptera litura), which suggested that CcPR-10 may be involved in cross-tolerance to abiotic and biotic stresses.89 The abundance of two PR-10 proteins from maize (ZmPR-10 and ZmPR-10.1) was increased by multiple abiotic stresses including SA, CuCl2, H2O2, coldness, darkness and wounding and biotic stresses such as Erwiniaste wartii and Aspergillus flavus infection.89
In vitro cryo protective activity was exhibited by PR-10 suggesting the role of some PR-10 proteins in frost-tolerance mechanisms.90 Another PR-10 homolog i.e. vegetative storage protein (VSP) from white clover (Trifolium repens L.), also accumulates under autumn and winter conditions, and thus may endow the plants with tolerance to chilling.91 Moreover, PR-10 proteins are over expressed in Oxytopis (Fabaceae) species adapted to the Arctic as opposed to temperate species.92 In a study by Vaas et al.,93 overexpression of PR-10a in suspension cultures of Solanum tuberosum causes an enhanced osmotic tolerance, which in turn leads to enhanced ability for cryo preservation. Abiotic stress-induced Zea mays PR-10 genes (ZmPR-10 and ZmPR-10.1) were also up-regulated following infection with pathogenic bacteria Erwinia stewartii and fungus Aspergillus flavus in young maize leaves and immature kernels, respectively.9
PR-10 proteins: A resource for crop improvement
The development of resistant cultivars with high yields and excellent quality is the most efficient, cost effective and environment friendly approach to prevent the losses caused by abiotic and biotic stress. Although some plants have remarkable ability to cope with extreme environmental onslaughts, however these stresses nevertheless represent a primary cause of crop-loss worldwide. Understanding the molecular process regulating these metabolic adaptations and untangling the network of interconnected signal pathways are important for developing stress resistant plants. Figure 3 displays different methods which can be applied to develop plants using PR-10 mediated resistance. An approach to transfer PR-10 mediated resistance in commercial cultivars is use of classical methods of plant breeding.
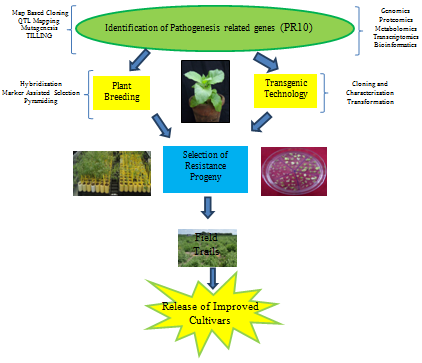
The steps needed for developing stress tolerant plants through traditional breeding approach are
However, the experiments needed here are very time consuming and laborious. Recent developments in genomics have potential to facilitate engineering for stress tolerance in plants.94 Advances in high-throughput sequencing and phenol typing platforms have potential to transform conventional breeding to genomics-assisted breeding and will address the challenge of increasing food yield, quality and stability of production through advanced breeding techniques. Next generation sequencing can help in the identification of the numerous PR-10 gene family members in the plant genome, and in the characterization of the associations with resistant phenotypes.
However, exploitation of the increasing knowledge of PR-10 proteins to enhance abiotic and biotic stress tolerance in the field should be exercised with caution. Sequence similarity of PR-10 with known allergens is a major setback in this area.3,39 Another less unexplored area is that manipulation of a PR-10 proteins might increase resistance to one pathogen or pest, but as an unwanted side effect might increase susceptibility to other pathogens or pests since induction or silencing of PR10 may affect the expression of other defense relates genes.36 Transgenic technologies have enormous potential to improve important crops by introduction of gene of interest often by Agrobacterium-mediated transformation or direct DNA transfer by particle bombardment method. Characterization of PR-10 proteins and development of transgenic plants overexpressing PR-10 proteins is important step in this direction. Table 1 lists the PR-10 genes which have been used to develop transgenic plants in different crop species.95–110 Multi-location field trials of transgenic plants expressing PR-10 will likely be next step for further evaluation.
Transgenic Plant |
Source of Transgene |
Host Resistance |
Gene Symbol |
Reference |
Tobacco |
Asparagus officinalis |
Oxidative stress |
AoPR1 |
|
Arabidopsis |
A. officinalis |
Oxidative stress |
AoPR10 (AoPR1) |
|
Arabidopsis |
Pinus monticola |
Cronartium ribicola, Wounding |
PmPR10-1.13 |
|
Brassica napus |
Pea |
Salinity |
PR 10.1 |
|
Arabidopsis |
Pea |
Salinity, cold and heat |
ABR17 |
|
Maize |
Maize |
Aspergillus flavus and Aflatoxins |
PR10 |
|
Faba bean |
Potato |
Drought and Salt |
PR10a |
|
Tobacco |
Peanut |
Salt and drought |
AhSIPR10 |
|
Arabidopsis |
Western white pine |
Cold |
PmPR10-1.10 |
|
Rice |
Soybean |
Salt |
GmPR10 |
|
Arachis hypogaea |
Arachis hypogaea |
A. flavus |
ARAhPR10 |
|
Arabidopsis |
Maize |
A. flavus, Pseudomonas syringae |
ZmPR10, ZmPR10.1 |
|
Tobacco |
Panax ginseng |
Colletotrichum gloeosporioides, Alternaria solani, SA, H2O2, JA, ABA, salt |
PgPR10-2 |
|
Potato |
Potato |
Salinity, Osmotic stress |
PR-10a |
|
Nicotiana benthamiana |
Alfalfa |
Wounding |
MsPR10.1A |
|
Vitis vinifera |
Vitis pseudoreticulata |
Plasmopara viticola |
VpPR10.2 |
|
Arabidopsis |
Pepper |
P. syringaepv. tomatoand Hyaloperonospora arabidopsidis |
PR-10/LRR1 |
|
Arabidopsis |
Panax ginseng |
Salt stress |
PgPR10 |
|
Soybean |
Soybean, tobacco |
Phytophthora sojae |
GmPR10 |
|
Soybean |
Soybean |
Phytophthora sojae |
Gly m 41 |
|
Banana |
A. hypogaea |
Salt and Drought |
AhSIPR10 |
Table 1 List of transgenic plants overexpressing PR-10 proteins for developing stress tolerant plants SA: Salicylic Acid; JA: Jasmonic Acid; ABA: Abscisic Acid
Our global food supply is threatened by multitude of abiotic and biotic stresses and advance molecular research techniques are trying to fill the gaps through understanding of plant resistance mechanism. PR-10 proteins are induced in response to pathogen and abiotic stimuli. Despite widespread reports on PR-10 involvement in combating a stress conditions sensed by plants, their functional mechanism is still unclear. However, many successful attempts were made to show the role of PR-10 proteins in stress resistance mechanism through transgenic approach in many species. Given the importance of the PR-10 proteins for abiotic and biotic stress tolerance, better understanding of these metabolic pathways involving PR-10 gene will be an exciting and rewarding process for plant scientists in the years to come.
The financial support received from North harvest Bean Growers Association is gratefully acknowledged. The authors thank Dr. Berlin Nelson, North Dakota State University for their helpful suggestions in preparing this article.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.