Advances in
eISSN: 2373-6402


Fusarium graminearum is responsible for Fusarium head blight (FHB), which is a destructive disease of wheat that accumulates mycotoxin such as Deoxynivalenol (DON) and makes its quality unsuitable for end use. Several FHB resistant varieties development is going on world-wide. However, the complete understanding of wheat defense response, pathogen (Fusarium graminearum) disease development mechanism and the gene crosstalk between organisms is still unclear. In our study focused to analyze pathogen (F. graminearum) molecular action in different Fusarium head blight resistance cultivars during the disease development. To understand the Fusarium graminearum pathogen molecular reaction, microarray gene expression analysis was carried out by using Fusarium graminearum (8x15k) Agilent arrays at two time points (3 and 7 days after infection) on three wheat genotypes (Japanese landrace cv. Nobeokabouzu-komugi - highly resistant, Chinese cv. Sumai 3-resistant and Australian cv. Gamenya - susceptible), which spikes infected byFusariumgraminearum‘H-3’strain. During the disease development the pathogen biomass as well as the expression of Trichothecene biosynthesis involved genes (Tri genes) in three wheat cultivars was determined. In our material no relation between fungus biomass and the disease symptoms were observed. However, it showed relation with fungus virulence factors expression (Tri genes). For the first time, we report the nature of Fusarium graminearum gene expression in the FHB-highly resistant cv. Nobeokabouzu-komugi during the disease development stage and the possible underlying molecular response.
Keywords: fusarium graminearum, fusarium head blight, wheat, trichothecene, microarray
FHB, fusarium head blight; DON, deoxynivalenol; NIV. nivalenol; GEO, gene expression omnibus; NCBI, national center of biotechnology information; Dai, days after inoculation
Fusarium head blight (FHB) is a destructive disease in wheat caused by Fusarium graminearum (F. g), which infects at flowering stage (anthesis) favored by warm and high humid climate. One of the main problems is accumulation of mycotoxin such as Deoxynivalenol (DON) and Nivalenol (NIV) during fungal spread and development in the grains. DON is the most prevalent mycotoxin in cereals.1 NIV chemo types are more restricted;2 therefore, more attention is generally focused on DON than on NIV as a trichothecene mycotoxin. Trichothecene mycotoxins are a large family of sesquiterpenoid secondary metabolites of Fusarium species. The trichothecene biosynthesis genes (Tri genes) are responsible for the mycotoxins biosynthesis. The current knowledge about trichothecene biosynthesis by F. g is derived from the core or main 25-kb region of the Tri-cluster. Among these clustered genes, Tri4, Tri5, Tri6 and Tri10 encode key factors for trichothecene biosynthesis pathway. Tri5 encodes Trichodiene synthase, which catalyzes the first committed step in the pathway.3 Tri4 encodes a cytochrome P450 mono-oxygenase involved in four oxygenation steps of the pathway after Trichodiene synthesis by Tri5.4 Tri genes are positively regulated by the transcription factors (TF) Tri65 and Tri10.6 This mycotoxin cause feed refusal or poor weight gain in animals and immunological problems in human.7 During fungal penetration and growth inside the plant, Fusarium proteases and mycotoxins act in a kind of strategic cooperation during spike and kernel colonization by featuring complementary roles during the host defense suppression and the intracellular colonization of spikelets. Gene expression studies are vital to understand the gene crosstalk between organisms to know the disease establishment and defense mechanisms. Here, we focused to analyze Fusarium graminearum molecular activity during the FHB infection.
Plant materials
The study was conducted using three wheat genotypes. Nobeokabouzu-komugi (Nobeokabouzu) is a Japanese landrace cultivar showing highly resistance to FHB and mycotoxin contamination.8 Sumai 3, a Chinese variety that possess Type II resistance9 and Gamenya, a highly susceptible variety.10 Seeds that were maintained as homozygous were taken from our laboratory for the study.
graminearum inoculation and sampling
Plants were grown in glass house in uniform condition. At early anthesis time of each spike, single floret inoculation with F. g strain ‘H-3’ which produces DON was carried out by pipette 10μl of the fungal suspension (1x105 macroconidia ml-1) between lemma and palea of each floret. At least 20 florets per spike were inoculated in a same day. Same time control plants were mock inoculated with 10μl of distilled water. The inoculated spikes were covered with a plastic bag for 72hours by avoiding any contact of bag and spike. Temperature and moisture content in the glass house were maintained at 25°C and 50% respectively. For RNA extraction, six spikes per genotype/treatment/time point were sampled. The time points are 0, 3 and 7days after inoculation (dai). Samples were immediately frozen in liquid nitrogen and stored at -80°C. For the microarray analysis, samples of three biological replication were taken at 3 and 7dai.
RNA extraction cRNA synthesis
Two infected spikes/genotype/replication were collected for RNA extraction. In total three biological replications were made. Total RNA was extracted from collected samples by using Nucleo Spin RNA plant kit (Macherey-Nagel, Germany) then converted to cRNA labeled using Low Input Quick Amp Labeling kit (Agilent technologies) and fluorophore cy3-CTP. The samples were cleaned using RNeasy MinElute Clean up kit (Qiagen). RNA quantity and quality evaluation were conducted using ND-1000 spectrophotometer (Nano Drop) measurement and 1.5 % agarose gel electrophoresis.
Microarray assay
A F. g 8x15k v 1.0 array (Agilent Technologies, Germany) was used for the microarray analysis. The platform of the Gene Expression Omnibus (GEO) for wheat array and F. g array are GPL19869 on the National Center of Biotechnology Information (NCBI). Microarray assay was used to measure the changes in global gene expression among the three genotypes with and without (mock control) FHB infection at 3 and 7dai. cRNA labeling with one color, hybridization and further washing was carried out according to the Agilent technical protocol. In total, 12 samples were hybridized and biologically replicated three times (12 samplesx3 replication=36 samples). Gene intensities were extracted from the scanned images, and the data were analyzed using the Gene Spring 12.6 software (Agilent Technologies). The F. graminearum microarray data were deposited in the NCBI Gene Expression Omnibus (GEO) database GSE66772
Data analyses
Microarray data analysis was carried out in a systematic manner. After normalization and statistical analysis Benjamin Hochberg FDR, 2 ways ANOVA p (treatment type and treatment time point) cut-off of 0.05 and a fold change ≥2, the data were grouped by Venn diagram to categorize the genes. T-test was used for comparing the expression of Tri5 and Tri6 among the genotypes as well as the time points
Gene expression confirmation
The 0, 3 and 7days after inoculation cDNA samples and Brilliant III Ultra-Fast SYBR Green QPCR Master Mix kit (Agilent Technologies) were used for the expression confirmation. Each reaction contained 10μl of 2x SYBR Green Master Mix, 2μl of 100ng/μlcDNA, and 1μl of 20ρM forward and reverse primer in a final volume of 20 μl. The PCR reaction was carried out by using Mx Pro3000 Real-time PCR (Stratagene). For F. g biomass and for Tri5 and Tri6, an initial denaturation step of 95°C for 10min (segment 1), followed by 40 cycles of 95°C for 30 s, 65°C for 1min and 72°C for 30s (segment 2). The expression of target gene was normalized to the reference gene, γ-actin, and relative to the untreated mock infected control samples. The Tri12 gene expression was confirmed by following the method reported by Ward et al.11 The primers used for Tri genes expression analysis were designed using Quant Primer software12 and were based on published gene sequences (National Center of Biotechnology Information-NCBI) (Table 1).
Gene |
Sequence (5´→3´ ) |
Tri 5 |
F-GAGCAGTACAACTTTGGAGG |
R-ACCATCCAGTTCTCCATCTG |
|
Tri 6 |
F-ACAGTCTTTGAAAGCGGACG |
R-GGTCTTGAGCTGACTGAGG |
|
Tri 12 |
12CON-CATGAGCATGGTGATGTC |
12NF-TCTCCTCGTTGTATCTGG |
|
12-15F-TACAGCGGTCGCAACTTC |
|
12-3F-CTTTGGCAAGCCCGTGCA |
|
γ-actin |
R-TCTGTCCTTGGTCTTGAGAG |
F-AGATCCTTTCGGACATCGAC |
Table 1 Details of primers used for qPCR analysis
F. graminearumgeneγ-actin is control used for normalization.
In our recent study the host wheat F. g response genes and their relation with the disease symptoms was clarified13 and in this study we focused to analyze pathogen (F.g) molecular response differences among FHB highly resistant, resistant and susceptible wheat cultivars during the infection.
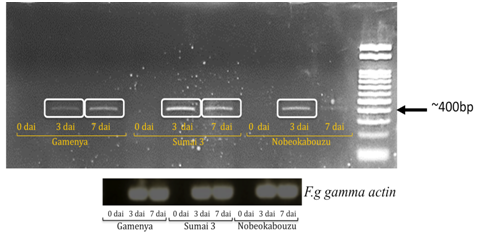
Expression of F. g γ-actin was determined in order to visualize pathogen development differences between the resistant and susceptible wheat cultivars
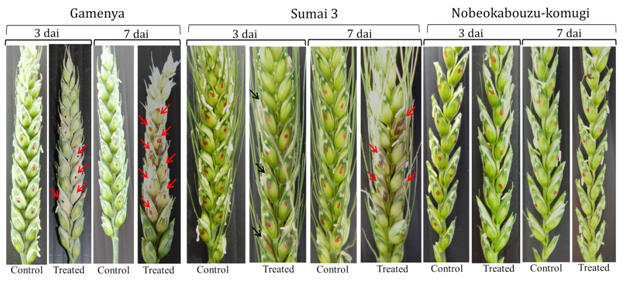
In FHB susceptible cultivar Gamenya, fungal biomass increased at 7dai, whereas in resistant cultivar Sumai 3 it was reduced (Figure 1). We expected minimum F. g biomass in highly resistant cultivar Nobeokabouzu-komugi, however, it increased drastically at 7dai. We assume that in susceptible cv. Gamenya, fungus biomass is less due to the less availability of nutrients required for the pathogen development when compare to resistant cultivars. Another reason might be the less requirement of fungus activity to infect susceptible cultivar necrotized tissue (Figure 2), however, in resistant cultivars (symptomless) required abundant fungus in order to suppress host defense genes, colonize and develop the disease. Here hypothesized that the F. g biomass does not have direct relation with fungal disease symptom, and depends on pathogen virulence as well as expression of virulence genes. In order to clarify the relation between fungus virulence gene expression and disease symptoms, expression of trichothecene biosynthesis genes (Tri genes) such as Tri5, Tri6 and Tri12 in both time points was determined.

Tri genes expression in FHB susceptible and resistant wheat cultivars
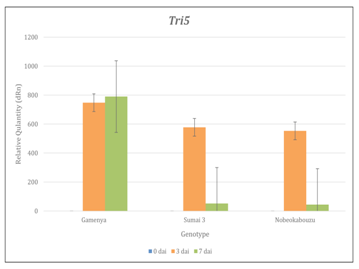
Tri genes expression was determined by using real time-PCR and RT-PCR and confirmed the expression of Tri5, Tri6 and Tri12 among three genotypes. The comparison of genotypes and time points showed that, in Gamenya, the expression of Tri5 was increased significantly at 7dai (t-test, p<0.05), where as in Sumai 3 and Nobeokabouzu reduced drastically (t-test, p<0.05) (Figure 3). This indicates that trichothecene biosynthesis is more abundant in FHB susceptible cultivar than in resistant cultivars. Also the peak of their biosynthesis in resistant cultivars identified at 3 dai, whereas in susceptible cultivar was at 7dai. In order to estimate the expression of others genes involved in trichothecene biosynthesis pathway, we determined the expression of transcription factor Tri6 (Figure 4). As expected, at 7dai in Gamenya their expression was increased significantly (t-test, p<0.05) and significantly reduced in the Nobeokabouzu and Sumai 3 (t-test, p<0.05). Also showed significant differences among the resistant genotypes (t-test, p<0.05). This result indicates the differences between resistant cultivars. F. g virulence during the infection stage depends on thrichothecene or mycotoxin release from the pathogen, for that reason we determined the expression of Tri12 by using RT-PCR (Figure 5). In Gamenya and Sumai 3 at both time points found their expression, whereas in Nobeokabouzu-komugi at 7dai there was no expression. This result demonstrated trichothecene biosynthesis and the product (mycotoxin) released in Gamenya and Sumai 3, but in Nobeokabouzu the trichothecene biosynthesis were reduced and the product released were not active at 7dai. One possibility is that the expressions of defense-related genes in this highly resistant cultivar are active that inhibit the trichothecene release and provoked toxicity to the fungus itself. However, in order to confirm this speculation it is important to quantify trichothecene (mycotoxin) content in these cultivars. In our laboratory previously determined the DON content in Gamenya, Sumai 3 and Nobeokabouzu at 10dai and the result clearly showed that DON content was high in Gamenya and less in Sumai 3 and Nobeokabouzu being 38.155, 13.500 and 10.955ppm, respectively.

Differentially expressed F. g transcripts in FHB susceptible and resistant cultivars
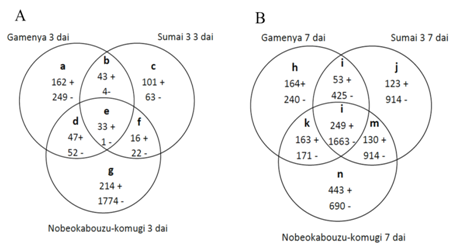
Another objective of this study was to identify differentially expressed F. g transcripts differences between FHB susceptible and resistant cultivars. By illustrating in Venn diagram, the F. g commonly expressed in three wheat genotypes and genes specifically expressed in each genotypes was determined (Figures 6) (Figure 7). Among wheat genotypes found that F. g up regulated genes is abundant in Nobeokabouzu-komugi, followed by Gamenya and Sumai 3. Also in Nobeokabouzu-komugi at 3dai is abundant the down-regulated genes, followed by Sumai 3 7dai, Nobeokabouzu-komugi 7dai, Gamenya 3dai, Gamenya 7dai and less in Sumai 3 3dai (Figure 6). We expected abundant F. g down regulated genes in highly resistant and resistant cultivars (Nobeokabouzu-komugi and Sumai 3) and less in susceptible cultivar Gamenya, however, the result showed opposite. One of the reason might be the rich availability of nutrients for the fungal growth in the resistant cultivar when compare to the susceptible cultivar with visible critical tissue damage such as necrosis (symptoms).
genotype-specific gene expression in highly resistant, resistant and susceptible wheat cultivars
With the objective to identify F. g genes specifically expressed in three wheat genotypes were analyzed by Venn diagram (Figure 7). In susceptible cultivar Gamenya, both up- and down-regulated gene expressions were not found significant differences among the time points (Figure 7A). In case of highly resistant Nobeokabouzu-komugi and resistant Sumai 3 found abundant up-regulated genes at 7dai whereas at 3dai found abundant down-regulated genes (Figure 7B) (Figure 7C). This showed concordance with wheat defense-related genes,11 which might avoid F. g activity at early stage of infection. The microarray study allowed identifying trichothecene biosynthesis involved genes and genes that were previously identified in wheat-F.g infection study. As expected in highly resistant Nobeokabouzu-komugi found down-regulation of Trichothecene biosynthesis involved genes, whereas in resistant Sumai 3 and susceptible Gamenya found only one down-regulated genes at 3 and 7dai respectively (Table 2).
Genotype |
Regulation |
Gene symbol or probe name |
Fold change |
Description |
Gamenya 3dai |
up |
FGSG_12985 |
4,1 |
Tri12; strain: NRRL13383 (NIV producer) |
Gamenya 7dai |
down |
FuGr_00017233 |
-8,3 |
Tri8; strain: H88-1 (NIV producer) |
down |
FGSG_03543 |
-6,7 |
Putative trichothecene biosynthesis gene |
|
Sumai3 3dai |
down |
FGSG_03528 |
-8,4 |
Conserved hypothetical protein |
Sumai3 7dai |
down |
FGSG_03533 |
-2,8 |
Related to TRI7 - trichothecene biosynthesis gene cluster |
Nobeokabouzu-komugi 3dai |
up |
FuGr_00017197 |
32,7 |
Tri4; strain: H88-1 (NIV producer) |
down |
FuGr_00017212 |
-8,1 |
Tri3; strain: H88-1 (NIV producer) |
|
down |
FuGr_00100141 |
-38,6 |
Tri3; strain: NRRL13383 (NIV producer) |
|
down |
FuGr_00017184 |
-17,9 |
Tri6; strain: H88-1 (NIV producer) |
|
down |
FGSG_03536 |
-19,4 |
Conserved hypothetical protein |
|
Nobeokabouzu-komugi 7dai |
down |
FGSG_03538 |
-2,5 |
Regulatory protein |
down |
FGSG_03529 |
-2,0 |
related to glucan 1,3-beta-glucosidase |
|
down |
FGSG_03530 |
-12,3 |
acetylesterase, trichothecene gene cluster |
|
down |
FGSG_03540 |
-28,5 |
Isotrichodermin C-15 hydroxylase |
|
up |
FuGr_00100130 |
7,2 |
Tri4; strain: NRRL13383 (NIV producer) |
|
down |
FuGr_00099876 |
-10,0 |
Tri12; strain: NRRL13383 (NIV producer) |
|
down |
FuGr_00099895 |
-9,6 |
Tri10; strain: NRRL13383 (NIV producer) |
Table 2 Transcriptomes involving in trichothecene biosynthesis
Differentially expressed trichothecene biosynthesis involving genes. List of transcriptomes expressed in our material. ˗ sign indicates down regulation. In bold indicates the genes, which previously reported to be involved in trichothecene biosynthesis.
Dai, days after inoculation
graminearum biomass and transcriptome abundance in wheat genotypes
g biomass and transcriptomes abundance were determined in order to analyze the relation with FHB symptoms in susceptible, resistant and highly resistant wheat cultivars. In our study, pathogen biomass was determined by measuring γ-actin. In our material F. g biomass does not have relation with FHB symptoms. In Nobeokabouzu found abundant F. g biomass as well as transcriptomes when compare to Sumai 3 and Gamenya. We expected high biomass and abundant transcriptomes in FHB susceptible cultivar Gamenya at late stage of infection and less in resistant cultivar Sumai 3 and Nobeokabouzu at early stage of infection. However the result showed abundant biomass as well as transcriptomes in resistant cultivars at early stage of infection and less in susceptible cultivar at both time points. One possible reason is that, due to reduced level of nutrients in the susceptible cultivar results in accumulation of empty fungal hyphae.14 Another possibility is that, as the susceptible cultivar becomes increasingly necrotic, where most of the host defense mechanisms are defeated, fewer fungal genes may be required in order to colonize and develop the disease.15 In our infected material clearly observed that, susceptible cultivar was strongly damaged and surrounding inoculated florets became necrotic whereas in case of resistant cultivar Sumai 3 observed small brownish necrosis and symptomless in highly resistant Nobeokabouzu. We hypostatized that; F. g genes that encode virulence factors have more influence to the FHB symptoms than F. g biomass. In order to clarify this point transcriptomic analysis was conducted by using microarray study.
As expected in Nobeokabouzu many trichothecene biosynthesis involved genes such as Tri3, Tri6, Tri10 and Tri12 were down regulated whereas in Sumai 3 and Gamenya found Tri7 and Tri8 down regulation respectively. In F.g, Tri3 is known to encode 15-O-Trichothecene acetyltransferase and is essential for C15 acetylation. Studies with Tri3 deletion mutant shows that acetylation at C15 is required for DON biosynthesis.16 Also Tri3 is crucial for intermediate metabolite calonectrin in order to synthesize mycotoxins such as 3ADON, 15A DON, DON and NIV. The down regulation of this gene definitively reduces mentioned mycotoxins biosynthesis. Here, most interesting down-regulated gene is Tri6. Because Tri6 is a transcription factor and regulates expression of genes involved in the synthesis of DON and T-2 toxin.17 Down regulation of this gene affects completely the trichothecene biosynthesis pathway by reducing the expression of crucial genes in this pathway. Another down regulated TF is Tri10. Tri10 acts upstream of the cluster-encoded transcription factor Tri6. Also is important the expression of both Tri6 and Tri10 for the expression of other trichothecene genes, and the genes for the primary metabolic pathway that precedes the trichothecene biosynthetic pathway. Study with disrupted Tri10 in Fusarium sporotrichioides abolished T-2 toxin production and dramatically decreased the transcript accumulation for four trichothecene genes (Tri4, Tri5, Tri6, and Tri101) and an apparent farnesyl pyrophosphate synthetase (Fpps) gene.18 The down regulation of both transcription factors produces huge reduction in the trichothecene biosynthesis. Another interesting point in Nobeokabouzu is the down regulation of Tri12 genes. The gene Tri12 encodes a predicted major facilitator super family protein suggested to play a role in export of trichothecene mycotoxins produced by Fusarium spp. In addition, Tri12 plays a role in self-protection and influences toxin production and virulence of the fungus in plant.19 It is however, unclear how the Tri12 protein (Tri12p) may influence trichothecene sensitivity and virulence of the wheat pathogen F.g. This result indicates that either by down regulation of essential two transcription factors (Tri6 and Tri10) that can avoid the mycotoxin synthesis or by down-regulating toxin efflux pump (Tri12) can be reduced the trichothecene damage to the host. Trichothecene biosynthesis genes down-regulated in resistant cultivar Sumai 3 was Tri7. Tri7 gene is responsible for C-4 acetylation, and known to be absent in DON-producing F. g strain but present in nivalenol (NIV) –producing strain.20 This indicates that in this cultivar the production of NIV was inhibited, but not DON. In case of Gamenya Tri8 gene was down regulated. Tri8 encodes an esterase responsible for removal of the C-3 acetyl group from trichothecenes as a final step in biosynthesis. Previous studies have shown that Tri101 encodes a C-3 transacetylase that acts as a self-protection or resistance factor during biosynthesis and that the presence of a free C-3 hydroxyl group is a key component of Fusarium trichothecene phytotoxicity. Since Tri8 encodes the esterase that removes the C-3 protecting group, it may be considered a toxicity factor.21 This result indicates that even FHB in susceptible cultivar some defense genes are expressing and might inhibiting the expression of Tri8.
In both resistant cultivars up regulation of Tri4 was found. Tri4 encodes a cytochrome P450 monooxygenase (CYP) for hydroxylation at C-2 of the first committed intermediate Trichodiene in the biosynthesis of trichothecenes.4 Especially highly resistant Nobeokabouzu was clearly symptomless and found less mycotoxin accumulation when compare to others cultivars in spite of Tri 4 gene up regulation. In susceptible cultivar Gamenya found up regulation of Tri12 gene. This indicates that mycotoxin produced by F. g was actively exported and accumulated in the kernel as data determined in another study.
This study demonstrated the F. g biomass, transcriptomes and virulence genes (Tri genes) expression differences among the highly resistant, resistant and susceptible wheat genotypes. The data clearly showed the relation between FHB symptoms and F. g virulence factors. Interestingly, in our material the FHB disease symptom does not have relation with F. g biomass. However, it showed relation with fungus virulence factors expression (Tri genes). In order to clarify the clear reason of abundant F. g transcriptomes in highly resistant cultivar Nobeokabouzu and less in susceptible cultivar Gamenya will require additional studies. The findings in nature of FHB highly resistant cultivar Nobeokabouzu and their molecular activity will contribute for better understanding and as additional information of FHB disease research. In future, the development and advance in F. g gene functional annotation will allow understand more clearly molecular activities and differences among the genotypes.
None.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.