MOJ
eISSN: 2374-6920


Research Article Volume 7 Issue 5
1Department of Agronomy, Zhejiang University, China
2Jiaxing Academy of Agricultural Sciences Institute, China
3Department of Horticulture, Zhejiang A&F University, China
Correspondence: Chunhai Shi, Agronomy Department, College of Agriculture and Biotechnology, Zhejiang University, Yuhangtang Road 866, Hangzhou 310058, PR. China, Tel +86-57188982691
Received: September 19, 2018 | Published: October 31, 2018
Citation: Weidong X, Shi C, Cao Z, Cheng F, et al. The analysis for alteration in starch biosynthesis metabolism in a japonica rice grain mutant which does not accumulate starch.MOJ Proteomics Bioinform. 2018;7(5):162-167. DOI: 10.15406/mojpb.2018.07.00252
Rice grain‒filling is an important agronomic trait that contributes greatly to grain weight. A grain mutant from the japonica cultivar Nipponbare by mutagenesis with ethyl methane sulfonate (EMS), which had no accumulation of starch granules in endosperm with a transparent liquid during grain filling stage, was used to analyze the caryopsis development and starch biosynthesis metabolism in present study. Measurement of soluble substances in the liquid of developing endosperm showed that there was remarkably higher soluble sugar content in this no starch mutant. Semi‒quantitative reverse transcription‒PCR (RT‒PCR) analysis of the starch‒synthesizing genes revealed that soluble starch synthase1 (SSS1) gene could be normally expressed in the mutant. Substantially lower expressions of starch branching enzyme1 (SBE1), isoamylase1 (ISA1) and pullulanase (PUL) were detected in the no starch mutant compared with the wild type, whereas the expression of ADP‒glucose pyrophosphorylase large subunit 1 (AGPL1) and ADP‒glucose pyrophosphorylase small subunit 1 (AGPS1) were visibly increased. The measurements of the key enzymatic activities involving in starch biosynthesis suggested that there was a significant reduction in the activities of starch debranching enzyme (DBE) and starch branching enzyme (SBE) in the transparent liquid endosperm of the no starch mutant, which coincided with their down‒regulation expression at transcriptional levels. These results suggest that the transparent liquid with no starch accumulation at the filling stage in the glume of the mutant was likely due to abnormal transcriptional and enzymatic activities related to the DBE and/or SBE genes loci mutant.
Keywords: no starch mutant, starch‒synthesizing enzymes, physiological metabolism, rice (oryza sativa l)
ADPL1, ADP‒glucose pyrophosphorylase large subunit 1; ADPS1, ADP‒glucose pyrophosphorylase small subunit 1; DBE, starch debranching enzyme; EMS, ethyl methane sulfonate; ISA1, isoamylase1; PUL, pullulanase; RT‒PCR, reverse transcription‒PCR; SBE, starch branching enzyme; SBE1, starch branching enzyme1; SSS1, soluble starch synthase1; DAP, days after pollination; WT, wild type
Rice (Oryza sativa. L) is one of the most important staple foods worldwide and the development of rice grain is abundantly related to growth of the endosperm. The endosperm is initiated by the fusion between a sperm nucleus and two polar nuclei during the double fertilization process and undergoes a series of coordinated cellular and metabolic events, including starch accumulation, cell death and starch granule packaging during the storage phase.1 Starch generally accounts for about 70% of the total dry weight in cereal grains and serves as a primary source of food with a wide range of industrial applications.2 In previous reports, many efforts have been made to elucidate the metabolic mechanisms underlying the starch biosynthesis pathway in developing endosperms, and some key enzymes, involving ADP‒glucose pyrophosphorylase (AGP), sucrose synthase (Susy), soluble starch synthase (SSS), granule‒bound starch synthase (GBSS), starch branching enzyme (SBE) and starch debranching enzyme (DBE) have been considered as necessities for the starch biosynthesis in cereal endosperm. In recent decades, a series of mutants related to deficient starch biosynthesis have been generated and provided new insights into the complex mechanism of starch biosynthesis in cereal endosperms. Several isoforms including SS I, SS II, SS III, SS IV and GBSS had specific functions in initiation, elongation, branching and debranching of transient or storage starch synthesis in source and sink tissues, respectively.3 Among all of these SS isoforms, SS I was highly and specifically expressed in rice endosperm and performs about 70% of total SS activities, and its activity was also higher than that in other isoforms.4 Two related forms of SBE (SBE I and SBE II) exist in cereal endosperm. Loss of SBE I isoform activity could limit the synthesis of starch in a way that cannot be compensated by SBE II isoform; thus, the activity of SBE I was essential for normal regular organization of the starch granule whether SBE II existed or not.5 Two DBE families occurred, such as ISA and PUL in plants.6 reported that sugary‒1 phenotype was caused by the loss of the activity of the PUL enzyme, suggesting that the DBE was also involved in starch biosynthesis apart from its function in starch degradation in conjunction with other hydrolytic activities Rice, barley and maize sugary mutants were generally caused by the lack of ISA genes and low DBE activity, accompanied with a decreased starch accumulation in the endosperm and alterations in the fine structure and numbers of starch granule.7 At least three ISA genes were present in plants. Among them, ISA 1 was highly expressed in the cereal endosperm and the reduced levels of ISA 1 could be found in the synthesis of phytoglycogen in the antisense‒expression of transgenic rice plants,8 which suggested that ISA 1 played a crucial role in proper starch biosynthesis. Down‒regulation of a PUL inhibitor activity in barley causes a reduction in the small (B‒type) granules, and also reduces amylose content, alters amylopectin glucan chain‒length distribution and reduces starch content.9 Moreover, the low activity of SBE 1 was also detectable for some Sugary mutants apart from the much lower DBE activity in endosperms, resulting from the lack of ISA genes.10,11 The visible reduction of SBE 1 activity in the flo2 mutant endosperm of rice implied that the activity of SBE 1 was partially responsible for the aberrant structure of storage starch.12 reported that the PUL gene encoding a pullulanase was highly expressed during the entire seed development period with a peak mid stage. It is possible that these enzymes do not function separately but constitute a larger enzyme complex, which could determine the fine structure of starch. These enzymes could cause the differences in abnormal metabolism of starch synthesis in RNA transcription and protein expression level.13 The results from some endosperm mutants, such as sugary, shrunken and floury revealed that the reduction of cytosolic ADP glucose pyrophosphorylase activity in shrunken endosperm could not inhibit granule initiation, but severely restrain the subsequent enlargement of granules, in which the shrunken endosperm often developed pleomorphic amyloplasts containing a large number of underdeveloped granules.14‒16 Moreover,17 documented that rice SPK, a Calmodulin‒like domain protein kinase, was required for storage starch accumulation via catalyzing sucrose synthase phosphorylation the substrate of which could be supplied for starch biosynthesis. However, all of these mutants mainly showed shrunken grains, lower weight or white‒core endosperm phenotypes. Little information was available on the mutant with the no starch accumulating phenotype in rice grains up to now. In present experiment, a complete no starch mutant isolated in grain from the japonica cultivar Nipponbare by mutagenesis with EMS was used to investigate its alteration in sugar cleavage and the starch biosynthetic pathway to confirm the regulatory system involved in the production of storage substances during the caryopsis development.
Plant materials
The no starch mutant, which had absolutely no accumulation of starch granules in the endosperm and showed an abnormal phenotype with the transparent liquid instead of starch granules during grain filling stage, was isolated from japonica rice cultivar Nipponbare (Oryza sativa L), by mutagenesis with EMS. Rice plants were grown and managed normally in the paddy field of Zhejiang University, Hangzhou, China, in 2010. At heading stage, the suitable panicles were tagged, and the rice grains were sampled about 15DAP and quickly frozen in liquid nitrogen. Prior to this experiment, the no starch mutant was identified in M3‒M5 to ensure the reliability of their genetic mutations of seeds in the transparent liquid and abnormal phenotype.
Assays of viable pollen
Pollen viability was assayed according to the method proposed.18 Ten anthers were removed from the spikelets before flowering. The panthers were crushed into a fine powder on a glass slide and stained with 10µl of 1% (v/v) of I2 in 1% (v/v) KI to observe the pollen viability using the light microscope. Pollen grains which were completely round in shape and stained entirely black were considered viable. The wild and mutant rice flowers were artificially pollinated by reciprocal crossing with hand emasculation. After two and a half hours, the pistils were removed and directly stained with aniline blue on a glass slide for a few minutes before observation by UV microscopy (LEICA DM IRB). Germinated and elongated pollen refers to the pollen grains attached to the stigma whose tube growth can be detected by UV fluorescence.
Measurement of soluble sugar contents in rice grains
Soluble sugar contents in rice grain of no starch mutant were determined by the method.19 A sample of 20‒dehulled rice were weighed and ground with mortar and pestle in 2ml of 0.7M perchloric acid and allowed to thaw to 4ºC on ice for 30min. The insoluble material was separated from the soluble metabolites including sugars by centrifugation at 12000g for 10min at 4ºC. The pellet was washed by re‒suspension followed by re‒centrifugation in 2ml of 0.7M perchloric acid and then the supernatant was neutralised with KOH/MES/KCl (2M KOH, 0.4M MES, 0.4M KCl), centrifuged at 2000g for 5min at 4ºC to remove insoluble potassium perchlorate and four 0.2‒ml replicate aliquots of the resulting supernatant were assayed for sugars. Sucrose was converted to glucose and fructose by the addition of 0.1ml 0.22M sodium acetate (pH 4.8) containing 3.7U invertase (β‒fructosidase) to two of the aliquots and incubated at 37ºC for 2h. The other two aliquots were incubated as above but with no invertase. Glucose and fructose were assayed in all 3 samples with standard spectrophotometric techniques employing hexokinase, glucose 6‒phosphate dehydrogenase (NADP‒dependent) and phosphoglucose isomerase as coupling enzymes. Free glucose and fructose contents were measured in the undigested controls. To calculate the total soluble content and the sucrose, the glucose and fructose content in the undigested controls were subtracted from those in the digested samples.
Scanning electron microscope analysis
Dehulled rice samples were pre‒fixed with 2.5% (v/v) glutaraldehyde in phosphate buffer (pH 7.0) for more than 4h, rinsed three times (15min each time) with phosphate buffer, and fixed overnight with 1% (v/v) OsO4 in phosphate buffer at 4ºC. They were then washed three times (15min each time) by phosphate buffer, dehydrated through an ethanol series (50, 70, 80, 90, 95 and 100%) (v/v) for 15min at each step, incubated in a 1:1 (v/v) ethanol‒isoamyl acetate mixture for 30min and then transferred to pure isoamyl acetate for 1h. Finally, the samples were dried to critical point with liquid CO2, and coated with gold‒palladium before they were mounted for observation under a scanning electron microscope (SEM) (Hitachi TM‒1000 Tabletop Microscope).20
RNA isolation and RT‒PCR analysis
Total RNA was extracted from the WT and mutant caryopsis using RNA iso‒mate for plant tissue and RNAiso TM Plus (TaKaRa) according to.20 All extracted RNAs including SBE 1, ISA 1, SSS 1, PUL, ADPL 1 and ADPS 1 were treated with R Nase‒free DNaseI to eliminate genomic DNA contamination according to the protocols recommended by the manufacturer, and checked by formaldehyde denatured agarose electrophoresis and quantified by spectrophotometer analysis before storing at ‒80ºC. The first strand of cDNA was synthesized from 2µg of total RNA using the Reverse Transcriptase M‒MLV (TaKaRa). RT‒PCR was performed with pairs of starch‒synthesizing enzyme‒gene‒primers Table 1. The rice Actin1 gene specific primers (forward, 5’‒CGTCAGCAACTGGGATGATATG‒3’; reverse, 5’‒GTGTGGCTGACACCATCACCAG‒3’) were used as a control.
Gene Name |
Accession No |
Forward primer5′→ 3′) |
Reverse primer(5′→ 3′) |
Amplicon Size(bp) |
SBE1 |
AK065121 |
TGGCCATGGAAGAGTTGGC |
CAGAAGCAACTGCTCCACC |
191 |
ISA1 |
AB093426 |
TGCTCAGCTACTCCTCCATCATC |
AGGACCGCACAACTTCAACATA |
132 |
PUL |
AB012915 |
ACCTTTCTTCCATGCTGG |
CAAAGGTCTGAAAGATGGG |
202 |
SSS1 |
AK109458 |
GGGCCTTCATGGATCAACC |
CCGCTTCAAGCATCCTCATC |
279 |
ADPL1 |
AK100910 |
ATGCAGTGCAGTGCGTCTTT |
ACTTCACTCGGGGCAGCTTA |
183 |
ADPS1 |
AK073146 |
AGAATGCTCGTATTGGAGAAAATG |
GGCAGCATGGAATAAACCAC |
258 |
Table 1 The primers sequences of starch‒synthesizing enzymes genes involved in starch synthesis pathway
All the primers were quoted.18
Assay of starch‒synthesizing enzymes activity
To prepare the enzyme extract, twenty caryopsis were hand‒homogenized at 4ºC in a mortar and pestle with 5ml extraction buffer containing 100mM tricine‒NaOH (pH=7.5), 8mM MgCl2, 2mM EDTA, 12.5% (v/v) glycerol; 1% (w/v) PVP‒40, 50mM 2‒mercaptoethanol. After centrifugation at 10000g for 25min, the supernatant solution and deposition were collected separately and used as the preparation for individual enzyme analysis. DBE activity was measured according to. The enzyme was measured by monitoring the change in absorbance of NADPH at 340nm in an assay mixture (supernatant) of 200µl in BIO‒RAD Smart Spec TM Plus spectrophotometer (Bio Rad Laboratories, Hercules, CA, USA). ADPG‒Pase, SBE, SSS and phosphorylase were assayed as described.20
Morphological phenotypes
No visible differences in agronomic or morphological traits were observed at the vegetable stage between the no starch mutant and WT (Nipponbare). However, some significant differences were detected in the grains. The no starch mutant exhibited an abnormal phenotype with transparent liquid filling the developing grains instead of starch after flowering. The mutant was quite different from its WT in their grain phenotypes after the initial grain filling. The inner and outer glumes of grains in the WT were well closed together, whereas those in the mutant plant were mostly kept open, exposing the seed from the crack at 15DAP (Figure 1A). The dehulled rice in the WT were filled with the starch granules which were milky white color at 15DAP and gradually became hard at maturity, while those in the mutant were full of watery transparent liquid that could remain green in the pericarp until the late filling stage (Figure 1B). Otherwise, the mutant had no embryo in the caryopsis, and the caryopsis was less plump than of the WT at different filling stages (Figure 1C & Figure 1D). The pollen viability was also identified abnormal at the flowering time for the mutant plants (Figure 1E & Figure 1F). The mutant‒type pollens could not germinate or elongate neither on the mutant stigma nor on the wild‒type stigma (Figures 1G‒1J), while the pollens from the WT plant could normally germinate and elongate after artificial pollination (Figure lL). The wild‒type pollen could also germinate and elongate normally on the mutant‒type stigma (Figure 1M), and the pollen tubes were observed in the ovary at 2h after pollination (Figure 1N). The hybrid F1 seeds could not be obtained by crossing the wild‒type pollen to the mutant‒type. These results all indicated that the mutant pollens were sterile.
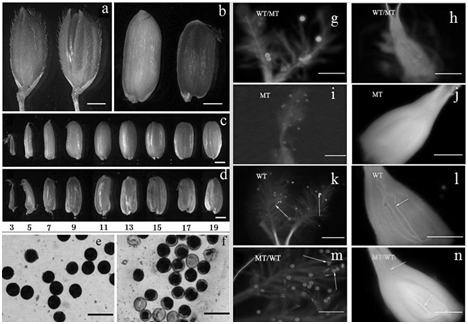
Figure 1 Morphological characterization of the wild type. a and b, rice grain at 15 DAP (a is the grain with glume, wild type in left and the mutant in right; b is the grain without glume, wild type in left and the mutant in right); c and d, whole seed at different filling time, (c is the wild type; d is the mutant) the numbers showed the days after flowering; e and f, pollen grains stained with I2‒KI solution (e was from wild type and f was from mutant), which the pollen grains staining complete black were judged as viable. g, i, k and m were 30min after artificial pollination and h, l, j and n were 2h after artificial pollination. g, h, k and l were the wild‒type stigmas, and i, j, m and n were the mutant‒type stigmas. Arrowheads show pollen tube elongation. Bars=1mm (A‒D) or 0.1mm (E‒N’).
Endosperm ultra‒structure
Distinctive differences of caryopsis under SEM between WT and the mutant were found at 15 DAP (Figure 2). SEM images of cross‒sectioning of the caryopsis in WT revealed that the endosperm was filled with densely packed, large and irregularly polyhedral starch granules, while there was no starch granules accumulation in the caryopsis, but rather an empty layer of pericarp for the mutant (Figure 2B). In addition, sucrose, fructose, glucose and total soluble sugar levels in the caryopsis of the mutant plant were significantly higher than those in WT (Figure 3), suggesting that the endosperm in the mutant had far less starch granules and cell tissue stocked with the increasing soluble sugar contents in its liquid endosperm.
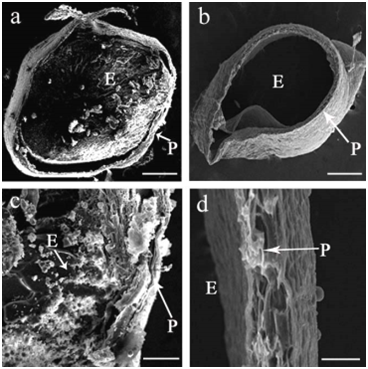
Figure 2 Scanning electron microscopic photographs of starch showing the cross‒section of the wild type (a, c) and the mutant (b, d) at 15DAP. Compound starch granules and the pericarp indicated by arrow, P, pericarp; E, endosperm; bars = 0.5mm (a, b) or 0.05 (c, d).
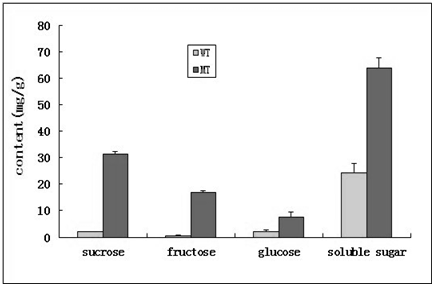
Figure 3 Sugar content levels in developing seeds at 15 DAP from the WT and the no starch mutant (MT). Bars represent the means of SD of three replicate samples.
Starch‒synthesizing enzymes
To elucidate the genotypic differences in sugar cleavage and starch biosynthesis in the developing caryopsis, the endosperm was sampled from the WT and mutant plants at 15 DAP and their mRNA levels were investigated by RT‒PCR. Significant differences in mRNA expression levels and temporal patterns of some genes related to starch biosynthesis except SSS1 were observed between the WT and mutant. Among them, transcriptional levels of SBE, ISA1 and PUL genes were considerably lower in the mutant than that in the wild‒type, whereas those of ADPL1 and ADPS1 were higher compared to those in the WT (Figure 4). SBE and DBE activities were also remarkably reduced in the mutant at the filling stage (Figure 4 & Figure 5).
Starch biosynthesis is a prerequisite process for the formation of normal endosperm in rice and other cereal grain fillings. It is difficult to form starch granule and to accumulate normal starch if this metabolic process does not work properly. AGPase is mainly located in the cytosol of cereal seed endosperm, accounting for about 85‒95% (w/v) of total AGPase activity in their endosperms.21 The rice shrunken mutant was caused by a lower AGPase activity in the filling endosperms.22 However, the decrease of cytosolic AG Pase activity in shrunken endosperm did not inhibit the granule initiation and only restrained the subsequent enlargement of granules severely after starch granule formation.23 SSS elongates glucans by adding Glc residues from ADP‒Glc to the glucan nonreducing ends through a‒1,4 linkages. However, the complete lack of SSS1, being a major SS isozyme in the developing endosperm, had been demonstrated to have little effect on the size and shape of starch granules and the crystallinity of endosperm starch, suggesting that other SS enzymes are probably capable of partly compensating SSSI function. In summary, this study strongly suggested that amylopectin chains are synthesized by the coordinated actions of SSI, SSIIa, and SSIIIa isoforms. In present study, the enlargement of the mutant ovary was not a result of double fertilization but due to parthenogenesis. The mutant with no starch in grain used in present experiment was quite different from the conventional grain mutants such as the sugary or OsAGP or shrunken described previously. The sugary mutant had seeds that shrank to a noticeable degree but did not contain detectable granules. Analysis of the OsAGP mutant revealed that a lesion of the two isoforms, OsAGPL2 and OsAGPS2b, caused a shrunken endosperm due to a remarkable reduction in starch synthesis.24 In contrast, the mutant presented here had a distinctive phenotype, and was found to have no embryo in its caryopsis and also no starch granule in filling endosperm. Moreover, this mutant had significantly sucrose, fructose, glucose and total soluble sugar contents in the caryopsis than in the WT, accompanied by an abnormal grain phenotype with watery transparent liquid in the developing grains instead of starch granule. In previous reports, many grain mutants with sugary or OsAGP or shrunken phenotypes were genetically caused by the deficiency of PUL or AGP function. However, the detectable starch granule and starch accumulation could be observed in the sugary or OsAGP or shrunken mutants, despite reduced grain weight and starch amount in their endosperms. In present study, there was no starch granule accumulation in the caryopses of the mutant. We had investigated the enzymatic activities and transcriptional expression of various genes related to starch biosynthesis pathway in the developmental endosperms for no starch mutant, this mutant displayed markedly lowered activities of DBE and SBE, and also the down‒regulating of transcripts of ISA1, PUL and SBE relative to its WT, which was similar to some sugary mutants. In previous studies, it had been revealed that DBE play an important role on the formation of highly organized clustered structure of phytoglycogen and starch granule. Rice sugary mutant exhibited the lowering activities of isoamylase‒type DBE, but granules were detectable in the rice endosperm. The lack of ISA in barely and maize had a profound effect on the structure, number and timing of initiation of starch granule,25‒30 had found that the lowering SBE activities in rice endosperms was concomitantly caused by the deficiency of DBE function for sugary mutant rice, suggesting that the lack of ISA1 had a profound impact on the activities of DBE and SBE in rice filling endosperms. Moreover, PUL was partially overlapped with ISA1 in the regulation of starch granule formation, although the deficiency of PUL had relatively smaller contribution to the aberrant structure of starch granule than that of ISA1. Hence, we deduced that the no starch mutant possibly was genetically caused by the deficiency of ISA1 gene, which also resulted in the reduction of transcripts of PUL and SBE1 genes and decreased enzymatic activities of DBE and SBE. On the other hand, the abnormal grain filling and aberrant phenotype in this no starch mutant was not attributed to the deletion of the AGP and SSS genes or the inactivation of the related enzyme activities (AGPase and SSS), but due to the expression level and the transcription degree of the genes. Furthermore, the remarkably higher level of sucrose in the developing grains of the no starch mutant could be explained by the possibility that the metabolic process of sugar cleavage was also affected by the lowering enzymatic activities of DBE and SBE in the fake endosperm of no starch mutant, which could result in the lowering of starch biosynthesis and a preferential transfer of glucose from sucrose to total soluble sugar during the grain filling period. Hence, it could be presumed that the low activities of SBE and DBE in the mutant endosperm were mostly attributed to down‒regulated transcription of the DBE and SBE genes, thereby leading to no accumulation of starch granules through starch biosynthesis in no starch mutant at the filling stage. These results also showed that this no starch mutant may be a valuable material for genetic and biochemical research on carbohydrate synthesis in the rice caryopsis.31
This work was supported by National Science and Technology Support Program (2011BAD35B02), Zhejiang Provincial Natural Science Foundation of China (Z3100089), the Science and Technology Office for Zhejiang Province (2012C12901‒2), Program for Innovative Research Team in Zhejiang Province (2010R50024‒5) and the Program for Innovative Research Team in University (IRT1185). We thank Dr. Alfred Quampah in revising the English of the manuscript.
Authors declare that there is no conflict of interests.

©2018 Weidong, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.