Journal of
eISSN: 2377-4312


Research Article Volume 3 Issue 2
1Bacteriology Department, Zagazig University, Egypt
2Bacteriology Department, Animl Health Research Institute, Egypt
Correspondence: Soumaya El-Shafii, AHRI, Nady ElSaid street, Dokki, Giza, Egypt, Tel 2 0114 571 3431
Received: December 04, 2015 | Published: February 15, 2016
Citation: Ammar AMA, El-Shafii SSA, AMO A, et al. Be detection of multidrug resistance genes in Pseudomonas Aeruginosa isolated from bovine mastitic milk. J Dairy Vet Anim Res. 2016;3(2):43-49. DOI: 10.15406/jdvar.2016.03.00071
A total of 270 milk samples were collected from private and governmental cow`s farms (200 samples from mastitic cows and 70 from apparently healthy cows). The samples were collected from different provinces (130 samples from Dakahlia, 66 samples from Sharkia and 74 samples from Damieta). Twenty isolates of P. Aeruginosa were isolated from 270 total milk samples in a percentage of 7.4. Damietta providence showed high percentage P. Aeruginosa isolates 8.1% followed by Sharkia providence and Dakhlia providence (7.6% and 6.9% respectively). Biochemical identification provided 16 distinct biotypes. Isolates that used galactose, mannose, manitol, displayed a green pigment and β hemolysis belonged to the more frequent biotype (80%). Most isolates showed multidrug resistance to antibiotic disc used (17 types) the rate of resistance ranged from 25%-100%. In Quinolone group, 85% of isolates were resistant to nalidixic acid and in flouroquinolne group, the resistance was ofloxacin enrofloxacin, followed by ciprofloxacin, and (95%, 85% and 80% respectively). In aminoglycosides group, 70% of isolates were resistance to neomycin followed by amikacin (65%) and gentamycin (50%). In polypeptides and sulphonamides groups, all isolates were resistance to colstin sulfate and sulfa methaxozoletrimethoprim, while in β-Lactams pencillin, macrolide, tetracycline and chloramphenicol groups, 95% of isolates were resistance to ampicillin, Erythromycin, tetracycline and chloramphenicol repectively. In lincomides group, 85% of isolates were resistance to both lincomycin and Clindamycin. On the other hand, in β-lactams (Cephalosporine) group, the resistance was only 30% to cefadroxil and 25% to and cephalothin.
The detection of β-lactams multidrug resistance genes using multiplex PCR in 20 isolates was observed where 10% of isolates (2 isolates) harbour LAT-1 to LAT4, CMY-2 to CMY-7, BIL-1 genes; 4 isolates (20%) harbour MIR-IT ATC-1 and all is dolates harbour DHA-1, DHA-3 genes. The Detection of flouroquinolone multidrug resistance genes in 20 P. Aeruginosa isolates using singplex PCR, where 6 isolates harbored ParC gene and all isolates harbored gyrA gene.
Pseudomonas aeruginosa is a highly relevant opportunistic pathogen that causes diseases in both animals and humans. It is typically found in soil, water, skin flora, and most man-made environments, since it requires minimal amounts of oxygen for growth, thereby allowing it to colonize a multitude of both natural and artificial environments.1
In most dairy herds, occurrence of pseudomonas mastitis is only sporadic, but occasionally it may be a serious herd problem. In surveys, overall incidence is not over 3%, with most reports showing less than 1% of udder infections caused by P. aeruginosa, as it is usually regarded as an opportunist, being relatively non-invasive and producing disease more often after injury of debilitating conditions, or it is secondary to other infectious agents. Use of common or non sterile teat cannulas for intramammary administration of antibiotics has been involved in the introduction and spread of pseudomonas mastitis. Access to ponds of stagnant water has been associated with some herd problems.2
P. aeruginosa produces a thick biofilm and due to its dense colonization, it is able to resist many antibiotics, disinfectants, as well as UV light and infected patients can therefore be very difficult to be treated. Another factor that contributes to P. aeruginosa resistance is its Gram-negative cell wall that is composed of three layers; the inner plasma membrane, peptidoglycan, and its outer membrane. P. aeruginosa bacterium is naturally resistant to many antibiotics due to the permeabiliity barrier afforded by its Gram-negative outer membrane. This high level of resistance in P. aeruginosa is dangerous to patient health. Moreover, pseudomonas maintains antibiotic resistance plasmids, R-factors and resistance transfer fragment (RTFs), and it is able to transfer these genes by horizontal gene transfer (HGT), mainly transduction and conjugation.3-4 The aim of this work is phenotypic and genotypic identification of quinolones resist P. aeruginosa isolated from mastitic cattle.
A total of 270 milk samples were collected from private and governmental cow`s farms (200 samples from mastitic cows and 70 from apparently healthy cows). The samples were collected from different provinces (130 samples from Dakahlia, 66 samples from Sharkia and 74 samples from Damieta). The distribution of collected milk samples are illustrated in Table 1.
Province |
Mastitic |
Apparently Healthy |
Total |
Dakhlia |
100 |
30 |
130 |
Sharkia |
50 |
16 |
66 |
Damieta |
50 |
24 |
74 |
Total |
200 |
70 |
270 |
Table 1 Numbers of samples collected from different provinces
All samples were submitted for bacteriological examination according to Cruickshank et al.,5 Suspected isolates were confirmed by a series of biochemical identification according to Quinn PJ et al.,6 using API20 NE. In vitro antibiogram sensitivity of P. Aeruginosa isolates to antimicrobial agents was performed according to NCCLS7using disc diffusion technique. Tetracycline (TE 30μg ), Ampicillin (AM 10μg), Neomycin (N 30μg), Erythromycin (E 10μg), Nalidixic acid (NA 30μg), Chloramphenicol (C 30μg), Sulfa/trimethoprim (SXT 25μg ), Cephalothin (KF 30μg ), Amikacin (KA 30μg), Clindamycin (DA 2μg), Colistin sulfate (CT 2μg), Gentamicin (CN 10μg), Lincomycin (L 2μg) and Ernofloxacin (Er 10μg). Extraction of plasmid DNA was applied according to Birnboim & Doly.8
Multiplex PCR protocol for detection of β-lactams resistance genes9
PCR was performed with a final volume of 50µl in 0.5ml thin-walled tubes. Each reaction contained 20mM Tris-HCl (pH 8.4); 50mM KCl; 0.2mM each deoxynucleoside triphosphate; 1.5mM MgCl2; 0.6µM primers CITMF, CITMR, DHAMF, and DHAMR; 0.5µM primers, EBCMF, and EBCMR (Table 2) and 1.25U of Taq DNA polymerase. Template DNA (2µl) was added to 48µl of the master mixture and then overlaid with mineral oil. The PCR program consisted of an initial denaturation step at 94°C for 3min, followed by 25 cycles of DNA denaturation at 94°C for 30s, primer annealing at 64°C for 30s, and primer extension at 72°C for 1min. After the last cycle, a final extension step at 72°C for 7min was added. Five-microliter aliquots of PCR product were analyzed by gel electrophoresis with 2% agarose (Bio-Rad, Hercules, Calif.). Gels were stained with ethidium bromide at10µg/ml and visualized by UV transillumination. A 100bp DNA ladder according to Sambrook et al.10
Target(s) |
Primer |
Sequence (5_ to 3_, as Synthesized) |
Expected Amplicon Size (bp) |
Nucleotide Position |
LAT-1 to LAT-4, CMY-2 |
CITMF |
TGG CCA GAA CTG ACA GGC AAA |
462 |
478–498 |
CITMR |
TTT CTC CTG AAC GTG GCT GGC |
939–919 |
||
DHA-1, DHA-2 |
DHAMF |
AAC TTT CAC AGG TGT GCT GGG T |
405 |
1244–1265 |
DHAMR |
CCG TAC GCA TAC TGG CTT TGC |
1648–1628 |
||
MIR-1T ACT-1 |
EBCMF |
TCG GTA AAG CCG ATG TTG CGG |
302 |
1115–1135 |
EBCMR |
CTT CCA CTG CGG CTG CCA GTT |
1416–1396 |
Table 2 Primer for β-lactams resistance genes
PCR cycles for amplifying the flouroquinolone antidrug resistance genes11
The PCR was performed in a 25μl reaction mixture in a thermal cycler. 100bp ladder was used as DNA molecular weight standards. The primers used are listed in Table 3 and the PCR cycles were illustrated in Table 3, the amplified PCR products were visualized using agarose gel electrophoresis.10
Primer |
Sequences |
gyrA (F) |
(5’TTA AAA TTT GTC ACG AAT ATG CC 3’ |
gyrA (R) |
5’ AAC GAT ACG CTC ACG ACC AGT 3’) |
parC (F) |
5’ AAA AAC TAC TCT ACA TTC TTT GAA AGG AG3’ |
parC (R) |
5’ CAG TTG GGT GGT CAA TCA TGT ACC 3’ |
Table 3 Primers for fluoroquinolone resistant gene
In the present study, Table 4 showed the occurrence of P. aeruginosa in bovine mastitis milk and apparently healthy milk samples, where 20 isolates of P. aeruginosa were isolated from 270 total milk samples in a percentage of 7.4. Damietta providence showed the highest percentage P. aeruginosa isolates 8.1% followed by Sharkia providence and Dakhlia providence (7.6% and 6.9% respectively). Identification of P. aeruginosa was based on the production of pigment, oxidase, glucose, arginine, nitrate and morphological characters on different media. Production of pigment was observed in 16 isolates from 20 isolates, most of them displayed the characteristic green color (80%).
Province |
Mastitis Milk |
Apparently Healthy Milk |
Total of Isolates |
|||
No |
% |
No |
% |
No |
% |
|
Dakhlia |
7/100 |
7 |
2/30 |
6.6 |
9 |
6.9 |
Sharkia |
4/50 |
8 |
1/16 |
6.25 |
5 |
7.6 |
Damietta |
4/50 |
8 |
2/24 |
8.3 |
6 |
8.1 |
Total |
15/200 |
7.5 |
5/70 |
7.1 |
20 |
7.4 |
Table 4 Occurrences ofP. aeruginosa in bovine mastitis milk and apparently healthy milk
All isolates were hemolytic on blood agar and most (18 isolates) demonstrated β hemolysis(90%). All isolates used carbohydrate for OF media. Most isolates were positive for galactose (19 isolates 95%) and only 2 isolates (10%) metabolized rhamonose 19 isolates were positive for mannose and manitol utilization and for urea hydrolysis 18 isolates were positive(90%). The results of these tests provided 16 distinct biotypes. Isolates that used galactose, mannose, manitol, displayed a green pigment and β hemolysis belonged to the more frequent biotype (80%) (Table 5).
Test |
Biotype1 |
Biotype2 |
||
No |
% |
No |
% |
|
Positive for β Hemolysis |
18 |
90 |
2 |
10 |
Positive OF Media |
18 |
90 |
2 |
10 |
Positive Galactose |
19 |
95 |
1 |
5 |
Positive for Mannose |
19 |
95 |
1 |
5 |
Positive for Urea |
18 |
90 |
2 |
10 |
Negative Rhamanose |
18 |
90 |
2 |
10 |
Pigment Production |
16 |
80 |
4 |
20 |
Manitol |
18 |
90 |
2 |
10 |
Table 5 Biotyping of isolated P. aeruginosa
Table 6 represents the antibiograms of P. aeruginosa isolates (20), where most isolates showed multidrug resistance to antibiotic disc used (17 types) the rate of resistance ranged from 25%-100%. In Quinolone group, 85% 0f isolates were resistant to nalidixic acid and in flouroquinolne group, the resistance was ofloxacin enrofloxacin, followed by ciprofloxacin, and (95%, 85% and 80% respectively).
Antibiotic Disc (Symbol) |
Sensitive Isolates |
Resistance Isolates |
||
No. |
% |
No. |
% |
|
Quinolone |
||||
Nalidixic acid (NA) |
3 |
15 |
17 |
85 |
Flouroquinolone |
||||
Enrofloxacin (EFX) |
2 |
10 |
18 |
95 |
Ofloxacin (OFX) |
4 |
20 |
14 |
90 |
Ciprofloxacin (CI) |
1 |
5 |
19 |
80 |
Aminoglycosides |
||||
Amikacin (KA) |
7 |
35 |
13 |
65 |
Neomycin (N) |
6 |
30 |
14 |
70 |
Gentamicin (CN) |
10 |
50 |
10 |
50 |
β-lactams (Cephalosporine) |
||||
Cefadroxil (CFR) |
14 |
70 |
6 |
30 |
Cephalothin (KF) |
15 |
75 |
5 |
25 |
β-lactams (Penicillin) |
||||
Ampicillin (AM) |
1 |
0 |
19 |
95 |
Polypeptides |
||||
Colistin Sulfate (CT) |
0 |
5 |
20 |
100 |
Sulfonamides |
||||
Sulfa Methaxozoletrimethoprim (STX) |
0 |
0 |
20 |
100 |
Macrolides |
||||
Erythromycin (E) |
1 |
5 |
19 |
95 |
Lincomides |
||||
Lincomycin (L) |
3 |
15 |
17 |
85 |
Clindamycin (DA) |
3 |
15 |
17 |
85 |
Tetracyclin |
||||
Tetracycline (TE) |
1 |
5 |
19 |
95 |
Phenicol |
||||
Cloramphenicol (C) |
1 |
5 |
19 |
95 |
Table 6 Rate of antibiogram of 20 isolates of P. aeruginosa
In aminoglycosides group, 70% of isolates were resistance to neomycin followed by amikacin (65%) and gentamycin (50%). In polypeptides and sulphonamides groups, all isolates were resistance to colstin sulfate and sulfa methaxozoletrimethoprim, while in β-lactams pencillin, macrolide, tetracycline and chloramphenicol groups, 95% of isolates were resistance to ampicillin, Erythromycin, tetracycline and chloramphenicol repectively. In lincomides group, 85% of isolates were resistance to both lincomycin and Clindamycin. On the other hand, in β-lactams (Cephalosporine) group, the resistance was only 30% to cefadroxil and 25% to and cephalothin.
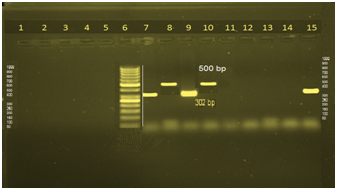
Table 7 showed the detection of β-lactams multidrug resistance genes using multiplex PCR in 20 isolates. 10% of isolates (2 isolates) harbour LAT-1 to LAT4, CMY-2 to CMY-7, BIL-1 genes; 4 isolates (20%) harbour MIR-IT ATC-1 and all is dolates harbour DHA-1, DHA-3 genes (Figure 1). Table 8 illustrated the Detection of flouroquinolone multidrug resistance genes in 20 P. aeruginosa isolates using singplex PCR, where 6 isolates harbored ParC gene and all isolates harbored gyrA gene (Figures 2 & 3).
Types |
bp of Fragments |
No. of Isolates (20) |
% |
LAT-1 to LAT-4, CMY-2 to CMY-7, BIL-1 |
462 |
2 |
15 |
DHA-1, DHA-2 |
405 |
0 |
35 |
MIR-1T ACT-1 |
302 |
4 |
20 |
Absence of β-lactams multidrug resistance genes |
No fragments |
14 |
15 |
Total |
20 |
100 |
|
Table 7 Detection of β-lactams multidrug resistance genes in 20 P. aeruginosa isolates using multiplex PCR
Types |
No. of Isolates (20) |
% |
parC |
6 |
30 |
Gyr |
14 |
70 |
Table 8 Detection of flouroquinolone multidrug resistance genes in 20 P. aeruginosa isolates using singplex PCR
Mastitis is probably the most important health disorder on dairy farms. This is reflected in relatively high incidence of clinical mastitis and, on many farms a high prevalence of subclinical mastitis. In most dairy herds, occurrence of pseudomonas mastitis is only sporadic, but occasionally it may be a serious herd problem. In surveys, overall incidence is not over 3%, with most reports showing less than 1% of udder infections caused by P. Aeruginosa, as it is usually regarded as an opportunist, being relatively non-invasive and producing disease more often after injury of debilitating conditions, or it is secondary to other infectious agents. Use of common or non sterile teat cannulas for intramammary administration of antibiotics has been involved in the introduction and spread of pseudomonas mastitis. Access to ponds of stagnant water has been associated with some herd problems.2
The present study was firstly aimed to isolate P. Aeruginosa from clinical mastitis and apparently healthy milk, where 20 isolates of P. Aeruginosa were isolated from 270 total milk samples in a percentage of 7.4. Damietta providence showed high percentage P. Aeruginosa isolates 8.1% followed by Sharkia providence and Dakhlia providence (7.6% and 6.9% respectively). These result revealed that the occurrence of P. aeruginosa differ from locality to another. Sharma & Sindhu12 isolated P. Aeruginosa in a percentage of 0.78%. These differences may be attributed to hygienic measure where the bacterium has been detected in contaminated wash hoses in milking parlors, in water, spray nozzle and in contaminated antibiotic preparation.13 Also the ability to survive in moist environments contributes greatly to this organism’s ubiquitous presence in water.14
According to Kiska,15 the rates of utilization of gelatos, mannose, manitol, rihamnose by P. Aeruginosa are (1%, 79%, 68% and 22% respectively), indicating that that those tests may be useful to distinguish among isolates. However in the present study, amour homogenous utilization of carbohydrates was observed than reported by Kiska DL15 and, on the basis of this result, P. Aeruginosa isolates were classified in the same group, a fact indicating the poor discriminatory power of biotyping. This result agree with de Freitas ALP16 who reported that phenotypic methods have been used to screen isolates which may be further typed by a more discriminatory tests.
Resistance to beta-lactam antibiotics is multi-factorial but is mediated mainly by inactivating enzymes called beta-lactamases. These enzymes cleave the amide bond of the beta-lactam ring causing antibiotic inactivation and are classified according to a structural17 and a functional classification.18
In the present study, the high rate of resistant isolates of Pseudomonas Aeruginosa to 17 antibiotics is worrisome. Despite improvements in antibiotic therapy Pseudomonas Aeruginosa is intrinsically resistant to a number of antimicrobial agents frequently including multiple classes of antimicrobial agents.16
This intrinsic resistance, P. Aeruginosa frequently develops acquired resistance either by mutation in chromosomally-encoded genes or by the horizontal gene transfers of antibiotic resistance determinants. Development of multidrug resistance by P. Aeruginosa isolates requires several different genetic events such as acquisition of different mutations and/or horizontal transfer of antibiotic resistance genes. Hypermutation helps the selection of mutation-driven antibiotic resistance in P. Aeruginosa strains producing chronic infections, but the clustering of several different antibiotic resistance genes in integrons favors the concerted acquisition of antibiotic resistance determinants. Some recent studies have concluded that phenotypic resistance associated to biofilm formation or to the emergence of small-colony variants may be important in the response of P. Aeruginosa populations to antibiotics treatment.19
In Quinolone group, 9% 0f isolates were resistant to nalidixic acid and in flouroquinolne group, the resistance was ofloxacin enrofloxacin, followed by ciprofloxacin, and (95%, 85% and 80% respectively). In aminoglycosides group, 70% of isolates were resistance to neomycin followed by amikacin (65%) and gentamycin (50%). In polypeptides and sulphonamides groups, all isolates were resistance to colstin sulfate and sulfa methaxozoletrimethoprim, while in β-lactams pencillin, macrolide, tetracycline and chloramphenicol groups, 95% of isolates were resistance to ampicillin, Erythromycin, tetracycline and chloramphenicol repectively.
In lincomides group, 85% of isolates were resistance to both lincomycin and Clindamycin. On the other hand, in β-lactams (Cephalosporine) group, the resistance was only 30% to cefadroxil and 25% to and cephalothin. These results agree with Saderi et al.,20 Polotto et al.,21 who reported that P. Aeruginosa isolates were multidrug resistant and were resistant to the agents in 2 or more of the antimicrobial categories: β-lactam antibiotics, and the fluoroquinolone. This was in harmony with the finding that the occurrence of plasmid mediated efflux pumps could be the factor contributing to fluoroquinolone and β-lactams categories resistance observed in this study, while tolerance to tetracycline could be correlated to the low permeability of bacterial outer membranes.22‒24
The differences in proportion of antibiotic resistance between the different locations wards investigated may be due to the application of different policies in treatment of mastitis or differences in the management of the dairy farms. This high resistance to antibiotics indicates the improper use of antibiotics, as shown in many studies.25 Another public health concern regarding Mastitis is antibiotic residues in milk due to extensive use of antibiotics in the treatment and control of the disease. Antibiotic residues in foods can lead to severe reactions in people allergic to antibiotics and, at low levels, can cause sensitization of normal individuals and development of antibiotic-resistant strains of bacteria.26
Also, Livermore27 reported that imprudent and often regular administration of antimicrobials has, however, compounded the problem by enriching for resistant bacteria populations at the expense of sensitive ones. Beta-lactam antibiotics are typically used to treat a broad spectrum of Gram-positive and Gram-negative bacteria. Beta-lactamases produced by Gram-negative organisms are usually secreted, especially when antibiotics are present in the environment. Beta-lactamases are enzymes18,28,29 produced by some bacteria that provide resistance to β-Lactam antibiotics like penicillins, and cephamycins.
β-lactamase provides antibiotic resistance by breaking the antibiotics structure. These antibiotics all have a common element in their molecular structure: a four-atom ring known as a β-Lactam. Through hydrolysis, the lactamase enzyme breaks the β-Lactam ring open, deactivating the molecule's antibacterial properties.
AmpC β-lactamases are clinically important cephalosporinases encoded on the chromosome of many gram negative bacteria, where they mediate resistance to cephalothin, cefazolin, cefoxitin, most penicillins, and β-lactamase inhibitor/βlactam combinations.30 In many bacteria, AmpC enzymes are inducible and can be expressed at high levels by mutation. Overexpression confers resistance to broad-spectrum cephalosporins. Transmissible plasmids have acquired genes for AmpC enzymes, which consequently can now appear in bacteria lacking or poorly expressing a chromosomal bla AmpC gene, such as E. Coli, K. Pneumoniae, P. Aeruginosa and P. Mirabilis.30
Resistance due to plasmid- mediated AmpC enzymes is less common than ESBL (Extended spectrum ß-lactamase) production in most parts of the world but may be both harder to detect and broader in spectrum. AmpC enzymes encoded by both chromosomal and plasmid genes are also evolving to hydrolyze broad-spectrum cephalosporins more efficiently.30 Plasmid-encoded β-lactamases active against cephalosporins and penicillins may provide a mechanism for β-lactam resistance.31,32 For newer stable P-lactam antibiotics, the rapid appearance o fβ-lactam resistance during drug therapy and frequent therapeutic failures related to the expansion of multiple resistances to β-lactam antibiotics are now common.31
Organisms overexpressing AmpC β-lactamases are a major clinical concern because these organisms are usually resistant to all the β-lactam drugs, except for cefepime, cefpirome, and the carbapenems.33,34 Constitutive overexpression of AmpC β-lactamases in gram-negative organisms occurs either by deregulation of the AmpC chromosomal gene or by acquisition of a transferable AmpC gene on a plasmid or other transferable element. The transferable AmpC gene products are commonly called plasmid-mediated AmpC β-lactamases.34‒36 Organisms that constitutively over express the chromosomal genes are collectively called derepressed mutants.
Twenty-nine different plasmid-mediated AmpC genes have been identified to date and have been deposited in GenBank.37 In the present study expressed only 6 of plasmid-mediated AmpC genes to detect the presence of LAT-1 to LAT-4, CMY-2 to CMY-7, BIL-1 (2 isolates) and MIR-1T ACT-1 enzymes (4 isolates) which confer resistance to cephalosporins.38
Fluoroquinolones are bactericidal, rapidly acting resistance using the antimicrobial drugs with wide spectrums. They are effective against many Gram negative bacterial pathogens in vitro.39 Their effect against Gram negative bacilli, including P. Aeruginosa, is one of their most important. However, fluoroquinolones resistance among P. Aeruginosa isolates has increased at an alarming rate due to its extensive use, which severely limits their usefulness.40 On the present study, 6 isolates of P. Aeruginosa were showing DNA fragments of parC and all isolates showing the fragments of gyrA. As the major resistance mechanism to fluoroquinolones in P. Aeruginosa involves modification of type II topoisomerases (DNA gyrase and topoisomerase IV).41‒44
It was concluded that the occurrence of P. Aeruginosa in mastitc milk differ from locality to another. On the basis of the result in this study, P. Aeruginosa isolates were classified in the same group, a fact indicating the poor discriminatory power of biotyping.
Despite improvements in antibiotic therapy Pseudomonas aeruginosa is intrinsically resistant to a number of antimicrobial agents frequently including multiple classes of antimicrobial agents. The differences in proportion of antibiotic resistance between the different locations wards investigated may be due to the application of different policies in treatment of mastitis or differences in the management of the dairy farms. This high resistance to antibiotics indicates the improper use of antibiotics.45
In most dairy herds, occurrence of pseudomonas mastitis is only sporadic, but occasionally it may be a serious herd problem. Unhygienic measure, as using of common or non sterile teat cannulas for intramammary administration of antibiotics has been involved in the introduction and spread of pseudomonas mastitis. Access to ponds of stagnant water has been associated with some herd problems.
None.
Author declares that there is no conflict of interest.

©2016 Ammar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.